The goal of ngboost is to provide an R interface for the Python package NGBoost.
“NGBoost is a method for probabilistic prediction with competitive state-of-the-art performance on a variety of datasets. NGBoost combines a multiparameter boosting algorithm with the natural gradient to efficiently estimate how parameters of the presumed outcome distribution vary with the observed features. NGBoost performs as well as existing methods for probabilistic regression but retains major advantages: NGBoost is flexible, scalable, and easy-to-use.” (From the paper, Duan, et at., 2019, see here)
The development version from GitHub with:
# install.packages("devtools")
devtools::install_github("Akai01/ngboost")A probabilistic regression example on the Boston housing dataset:
library(ngboost)
#> Loading required package: reticulate
data(Boston, package = "MASS")
dta <- rsample::initial_split(Boston)
train <- rsample::training(dta)
test <- rsample::testing(dta)
x_train = train[,1:13]
y_train = train[,14]
x_test = test[,1:13]
y_test = test[,14]
model <- NGBRegression$new(Dist = Dist("Exponential"),
Base = sklearner(),
Score = Scores("MLE"),
natural_gradient =TRUE,
n_estimators = 600,
learning_rate = 0.002,
minibatch_frac = 0.8,
col_sample = 0.9,
verbose = TRUE,
verbose_eval = 100,
tol = 1e-5)
model$fit(X = x_train, Y = y_train, X_val = x_test, Y_val = y_test)
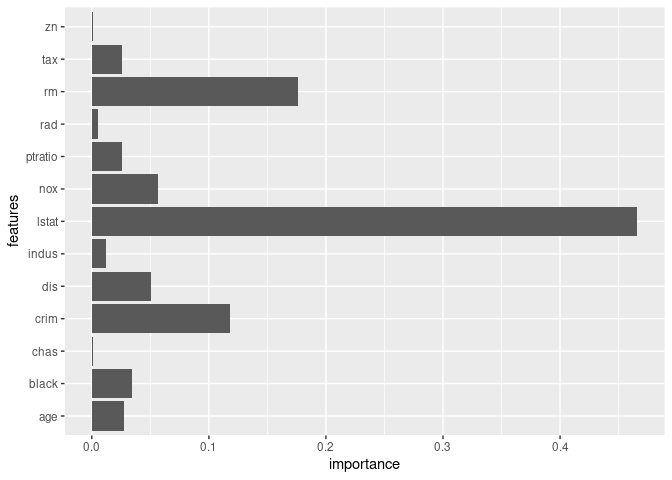
model$feature_importances()
#> features importance
#> 1 crim 0.117881241
#> 2 zn 0.001208810
#> 3 indus 0.012372080
#> 4 chas 0.001002271
#> 5 nox 0.056942928
#> 6 rm 0.176114767
#> 7 age 0.027487719
#> 8 dis 0.050332393
#> 9 rad 0.004942415
#> 10 tax 0.025936466
#> 11 ptratio 0.025421921
#> 12 black 0.034273781
#> 13 lstat 0.466083208
model$plot_feature_importance()model$predict(x_test)%>%head()
#> [1] 20.97577 20.26427 15.82709 15.25654 19.71702 14.75056
distt <- model$pred_dist(x_test) # it returns a NGBDist
class(distt)
#> [1] "NGBDistReg" "R6"
?NGBDist # see the available methods
#> No documentation for 'NGBDist' in specified packages and libraries:
#> you could try '??NGBDist'
distt$interval(confidence = .9)
#> [[1]]
#> [1] 1.0759162 1.0394212 0.8118237 0.7825583 1.0113507 0.7566050 1.0496568
#> [8] 1.1143298 1.0472284 0.9037815 0.9970665 1.0398567 1.5715081 0.9548589
#> [15] 1.0998399 1.1102689 1.1557172 1.1450350 1.6915874 1.6904072 1.1515997
#> [22] 1.1742950 1.2402832 1.4187055 1.2585190 0.9334092 1.0169164 0.9760542
#> [29] 1.0598535 1.0620905 0.8525441 1.0351985 0.8259550 0.9382414 0.8467941
#> [36] 0.8358114 0.8565694 0.8781342 1.0736043 0.8531474 1.2016832 2.3322218
#> [43] 2.2672122 1.1598534 1.0317973 1.2032399 1.7389629 1.4246417 2.1892182
#> [50] 1.6800488 1.1797544 1.0108459 0.9288902 0.9744566 1.1031933 1.0725771
#> [57] 2.1594903 1.3025033 1.1108417 1.0784915 1.1893328 1.2879130 1.3794029
#> [64] 1.7107134 1.1689120 2.2720635 1.7216025 1.0751543 1.2996765 2.1117497
#> [71] 1.5585570 2.2604965 2.1535752 1.1316002 1.1357918 1.3546891 1.6698481
#> [78] 1.3958033 1.4828655 1.4778899 1.0536363 1.0856019 1.0336042 1.1423934
#> [85] 1.1148042 1.1910388 1.1857402 1.2489614 1.0946772 0.9930221 0.9899008
#> [92] 1.0654861 1.0277351 1.6540368 1.4944613 1.5725638 1.1376886 1.1548331
#> [99] 1.6129248 1.7227833 0.5928467 0.5054178 0.7301362 0.8153189 0.3757723
#> [106] 0.6276727 0.6275357 0.3834950 0.8009085 0.7138012 0.4227620 0.5606350
#> [113] 0.8001861 0.6574251 0.6546425 0.8148326 0.8579452 0.8723704 1.0687165
#> [120] 1.1216865 0.7828301 1.0513031 1.0197157 1.1056430 1.0769412 1.0198085
#> [127] 1.5600706
#>
#> [[2]]
#> [1] 62.83778 60.70633 47.41373 45.70452 59.06690 44.18874 61.30413
#> [8] 65.08129 61.16230 52.78443 58.23265 60.73176 91.78232 55.76755
#> [15] 64.23502 64.84412 67.49848 66.87459 98.79543 98.72650 67.25800
#> [22] 68.58349 72.43747 82.85804 73.50251 54.51481 59.39196 57.00545
#> [29] 61.89966 62.03031 49.79196 60.45971 48.23906 54.79703 49.45614
#> [36] 48.81471 50.02706 51.28653 62.70276 49.82720 70.18308 136.21102
#> [43] 132.41420 67.74005 60.26107 70.27399 101.56234 83.20474 127.85905
#> [50] 98.12153 68.90235 59.03742 54.25088 56.91214 64.43088 62.64276
#> [57] 126.12282 76.07137 64.87757 62.98819 69.46176 75.21924 80.56261
#> [64] 99.91246 68.26911 132.69754 100.54843 62.79329 75.90627 123.33458
#> [71] 91.02592 132.02198 125.77735 66.08995 66.33475 79.11923 97.52577
#> [78] 81.52046 86.60524 86.31464 61.53655 63.40347 60.36659 66.72031
#> [85] 65.10900 69.56140 69.25194 72.94431 63.93350 57.99644 57.81414
#> [92] 62.22863 60.02382 96.60233 87.28248 91.84398 66.44553 67.44684
#> [99] 94.20122 100.61739 34.62460 29.51841 42.64285 47.61786 21.94659
#> [106] 36.65858 36.65058 22.39763 46.77624 41.68883 24.69098 32.74332
#> [113] 46.73405 38.39624 38.23373 47.58946 50.10741 50.94990 62.41729
#> [120] 65.51095 45.72039 61.40028 59.55545 64.57395 62.89765 59.56087
#> [127] 91.11432Classification example:
data(BreastCancer, package = "mlbench")
dta <- na.omit(BreastCancer)
dta <- rsample::initial_split(dta)
train <- rsample::training(dta)
test <- rsample::testing(dta)
x_train = train[,2:10]
y_train = as.integer(train[,11])
x_test = test[,2:10]
y_test = as.integer(test[,11])
model <- NGBClassifier$new(Dist = Dist("k_categorical", k = 3),
Base = sklearner(),
Score = Scores("LogScore"),
natural_gradient = TRUE,
n_estimators = 100,
tol = 1e-5,
random_state = NULL)
model$fit(x_train, y_train, X_val = x_test, Y_val = y_test)
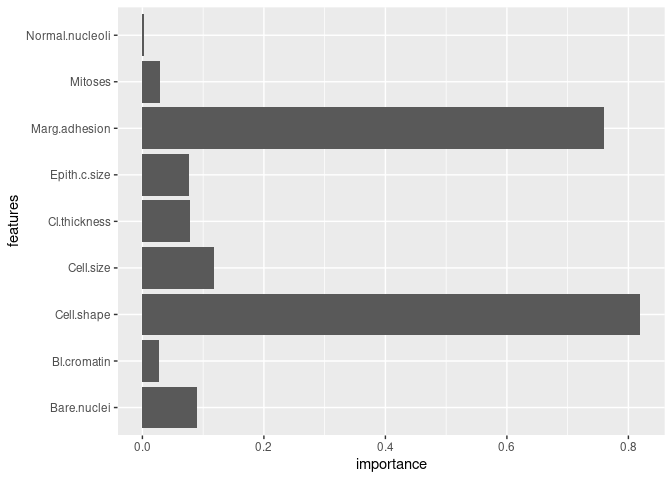
model$feature_importances()
#> features importance
#> 1 Cl.thickness 4.790004e-02
#> 2 Cell.size 5.227276e-02
#> 3 Cell.shape 7.486254e-01
#> 4 Marg.adhesion 7.475022e-01
#> 5 Epith.c.size 7.048404e-02
#> 6 Bare.nuclei 6.795669e-02
#> 7 Bl.cromatin 3.386698e-03
#> 8 Normal.nucleoli 2.263429e-03
#> 9 Mitoses 2.909698e-02
#> 10 Cl.thickness 3.022025e-02
#> 11 Cell.size 6.596945e-02
#> 12 Cell.shape 7.018171e-02
#> 13 Marg.adhesion 1.151351e-02
#> 14 Epith.c.size 5.616345e-03
#> 15 Bare.nuclei 2.190058e-02
#> 16 Bl.cromatin 2.398665e-02
#> 17 Normal.nucleoli 1.123269e-03
#> 18 Mitoses 1.004279e-10
model$plot_feature_importance()model$predict(x_test)
#> [1] 1 1 1 2 1 1 1 2 1 1 1 2 2 2 2 2 1 1 1 1 1 1 1 1 1 1 1 2 1 2 2 1 2 1 1 1 1
#> [38] 1 1 1 2 2 1 2 1 1 2 1 2 1 1 2 2 1 2 2 1 1 1 1 2 2 2 1 1 2 2 2 1 1 1 1 1 1
#> [75] 2 1 2 1 2 2 2 1 1 1 1 2 2 1 2 1 1 2 1 1 1 1 1 1 1 2 1 1 1 1 2 1 2 2 1 1 1
#> [112] 1 2 1 1 1 1 1 1 2 1 1 2 1 2 1 2 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 1 1 1 1 2 2
#> [149] 1 1 1 2 1 1 1 1 1 1 1 1 2 2 1 1 1 2 1 1 1 1 2
model$predict_proba(x_test)%>%head()
#> [,1] [,2] [,3]
#> [1,] 1.081404e-17 1.000000e+00 6.222232e-18
#> [2,] 1.081404e-17 1.000000e+00 6.222232e-18
#> [3,] 1.081404e-17 1.000000e+00 6.222232e-18
#> [4,] 1.081404e-17 6.222232e-18 1.000000e+00
#> [5,] 1.081404e-17 1.000000e+00 6.222232e-18
#> [6,] 1.081404e-17 1.000000e+00 6.222232e-18