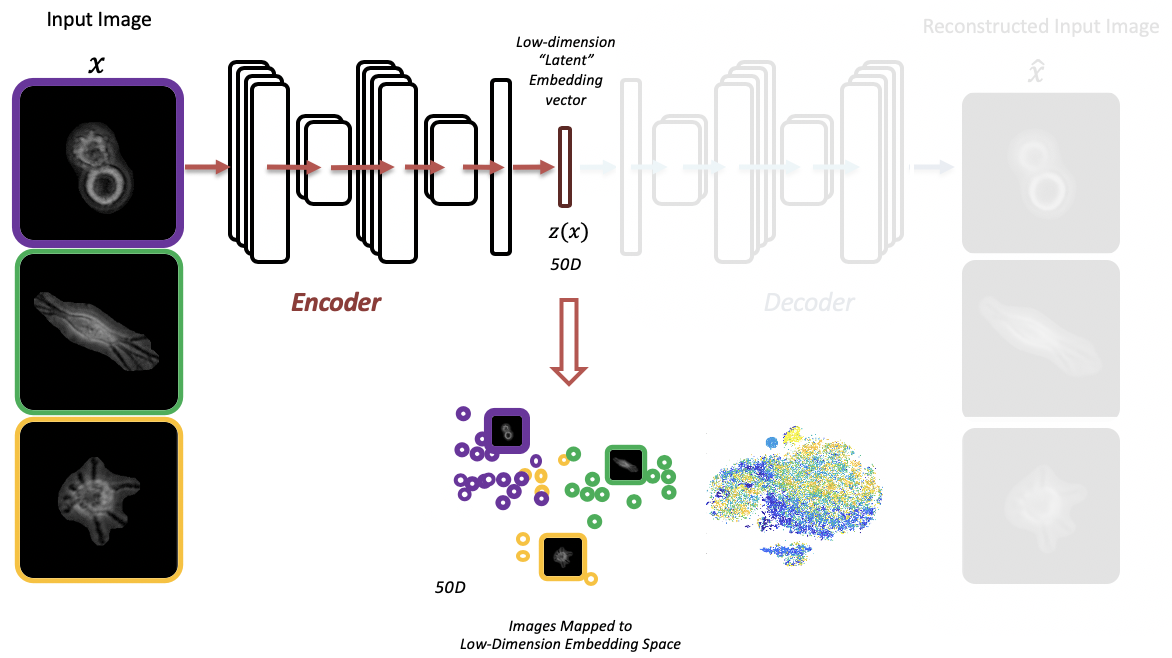
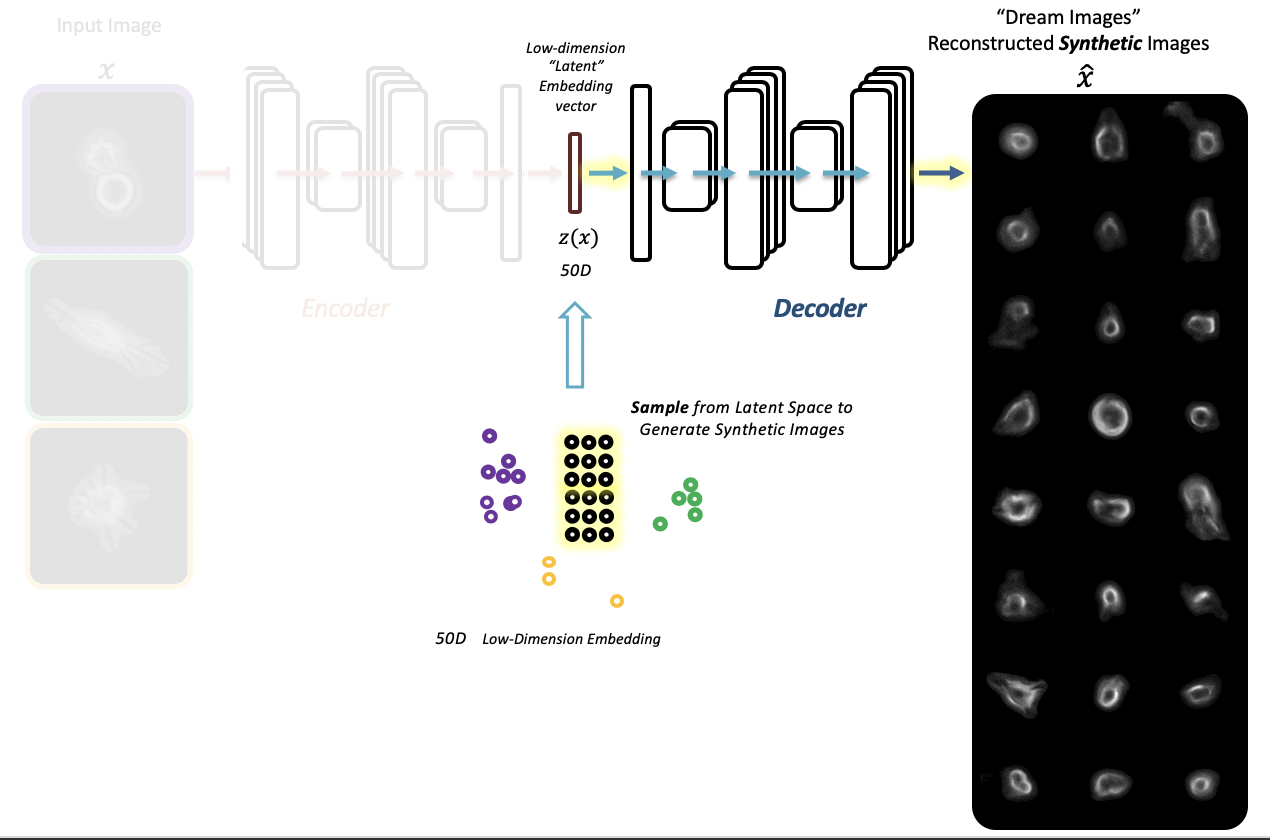
- Extracting latent features from label-free live cell images using Adversarial Autoencoders
- Interpretable deep learning uncovers cellular properties in label-free live cell images that are predictive of highly metastatic melanoma, Cell Systems, 2021, 12(7):733-747.e6, written by Assaf Zaritsky, Andrew R Jamieson, Erik S Welf, Andres Nevarez, Justin Cillay, Ugur Eskiocak, Brandi L Cantarel, Gaudenz Danuser.
Developed and tested on Red Hat Linux 7.
- also see more detailed information here
- Set-up compute environment with containers:
- Install Singularity
- Tested with Singularity 3.5.3
- Need CUDA 8.0+ compatible GPU and drivers (e.g. P100)
- Pull Singularity container .sif image file from Singularity Hub
- alternatively, a copy can be found here
- Definition file provided in this repo
singularity pull shub://andrewjUTSW/openLCH:latest- Test GPU
singularity exec --nv --cleanenv ./openLCH_latest.sif nvidia-smi
- Download 2000 image sample data set
curl https://cloud.biohpc.swmed.edu/index.php/s/FqZSqoKfHii6ony/download --output sample2000.tar.gz- unzip data
tar xvzf ./sample2000.tar.gz
- Included are a random sample set of 256x256 cell images
- Create image file list
ls `pwd`/data2/*.png > imagePathList.txt - Full data set provided here:
- Download previously trained autoencoder .t7 file
curl https://cloud.biohpc.swmed.edu/index.php/s/YAQQtpwTX2NKS89/download --output autoencoder_eval_56zTRAINED.t7
singularity exec --nv --cleanenv openLCH_latest.sif /bin/bash -c 'cd ./code; \
LCH_PATH=YOUR_CODE_PATH_HERE; \
th -i ./interp_LatentSpace_LCH_MD_single_2.lua \
-imPathFile $LCH_PATH/imagePathList.txt \
-autoencoder $LCH_PATH/autoencoder_eval_56zTRAINED.t7 \
-outDir $LCH_PATH/output/interpOut/ \
-gpu 1 \
-img1 51 \
-img2 81' singularity exec --nv --cleanenv openLCH_latest.sif /bin/bash -c 'cd ./code; \
LCH_PATH=YOUR_CODE_PATH_HERE; \
th ./run_mainLCH_AAE_Train_2.lua \
-modelname AAEconv_CLEAN \
-nLatentDims 56 \
-imsize 256 \
-imPathFile $LCH_PATH/imagePathList.txt \
-savedir $LCH_PATH/outputNew/ \
-epochs 100 \
-gpu 1' singularity exec --nv --cleanenv openLCH_latest.sif /bin/bash -c 'cd ./code; \
LCH_PATH=YOUR_CODE_PATH_HERE; \
th ./call_DynComputeEmbeddingsRobust_2.lua \
-autoencoder $LCH_PATH/outputNew/autoencoder_eval.t7 \
-imsize 256 \
-dataProvider DynDataProviderRobust_2 \
-imPathFile $LCH_PATH/imagePathList.txt \
-gpu 2 \
-embeddingFile $LCH_PATH/outputNew/embeddings_sampleTest.csv'Explore latent space by shifting embedding vector values (one dimension at a time) of an input cell image and reconstructing shifted synthetic cell images
singularity exec --nv openLCH_latest.sif /bin/bash -c 'cd ./code; \
LCH_PATH=YOUR_CODE_PATH_HERE; \
th -i ./exploreZ_LatentSpace_LCH_single_2.lua \
-imPathFile $LCH_PATH/imagePathList.txt \
-autoencoder $LCH_PATH/autoencoder_eval_56zTRAINED.t7 \
-outDir $LCH_PATH/outputOrig/zExploreOut \
-img1 10 \
-uR 1 \
-numSteps 6' singularity exec --nv --cleanenv openLCH_latest.sif /bin/bash -c 'cd ./code; \
LCH_PATH=YOUR_CODE_PATH_HERE; \
th -i ./zLatent2ReconBatchLCH_2.lua \
-autoencoder $LCH_PATH/autoencoder_eval_56zTRAINED.t7 \
-zLatentFile $LCH_PATH/outputNew/embeddings_sampleTest.csv \
-reconPath $LCH_PATH/outputNew/zRecon/ \
-nLatentDims 56'@article {Zaritskyj.cels.2021.05.003,
author = {Zaritsky, Assaf and Jamieson, Andrew R. and Welf, Erik S. and Nevarez, Andres and Cillay, Justin and Eskiocak, Ugur and Cantarel, Brandi L. and Danuser, Gaudenz},
title = {Interpretable deep learning uncovers cellular properties in label-free live cell images that are predictive of highly metastatic melanoma},
journal = {Cell Systems},
year = {2021},
volume = {12},
number = {7},
pages = {733-747.e6},
month = {Jul},
doi = {10.1016/j.cels.2021.05.003},
url = {https://doi.org/10.1016/j.cels.2021.05.003},
eprint = {https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8353662/}
}