PANAM2 is a pipeline for depicting the microbial community by implementing a phylogenetic approach for the annotation of the gene coding for 16S and 18S rRNA. It is based on PANAM (Taib et al. 2013) with some improvements and focusing on Illumina sequencing.
PANAM2 combines the publicly available tools: VSEARCH, HMMER, FASTTREE, KRONA, R (with packages Vegan, Phyloseq, Picante, Mass, Cluster) with 3 main perl scripts dedicated to:
-
step 1 - quality_panam.pl: merging, demultiplexing and cleaning raw Illumina reads
-
step 2 - preprocess_panam.pl: clustering and cleaning OTUs according to their abundances (remove rares for example)
-
step 3 - taxonomy_panam.pl: processing similarity annotations, alignments and phylogenies and computing main diversity indices for each samples and taxonomic units (system dependencies:bioperl)
and 2 optionnal scripts:
-
phylodiv_panam.pl (beta version): computing phylogenetic indices from detected clades or main taxonomic groups (MNND, NRI, NTI...) (system dependencies: R with packages Picante, Mass, Cluster)
-
postprocess_panam.pl: generating some additionnal richness indices, krona graphics, R data for processing additional analyses with phyloseq object and a HTML rapport displaying the main results from the previous scripts: (partial system dependencies: R with packages Vegan and phyloseq)
The results of these differents steps are writen in the following directoris: quality_output, preprocess_output, panam_output, PhyloDiv_output
The script panam2.pl allows to launch all the scripts described above
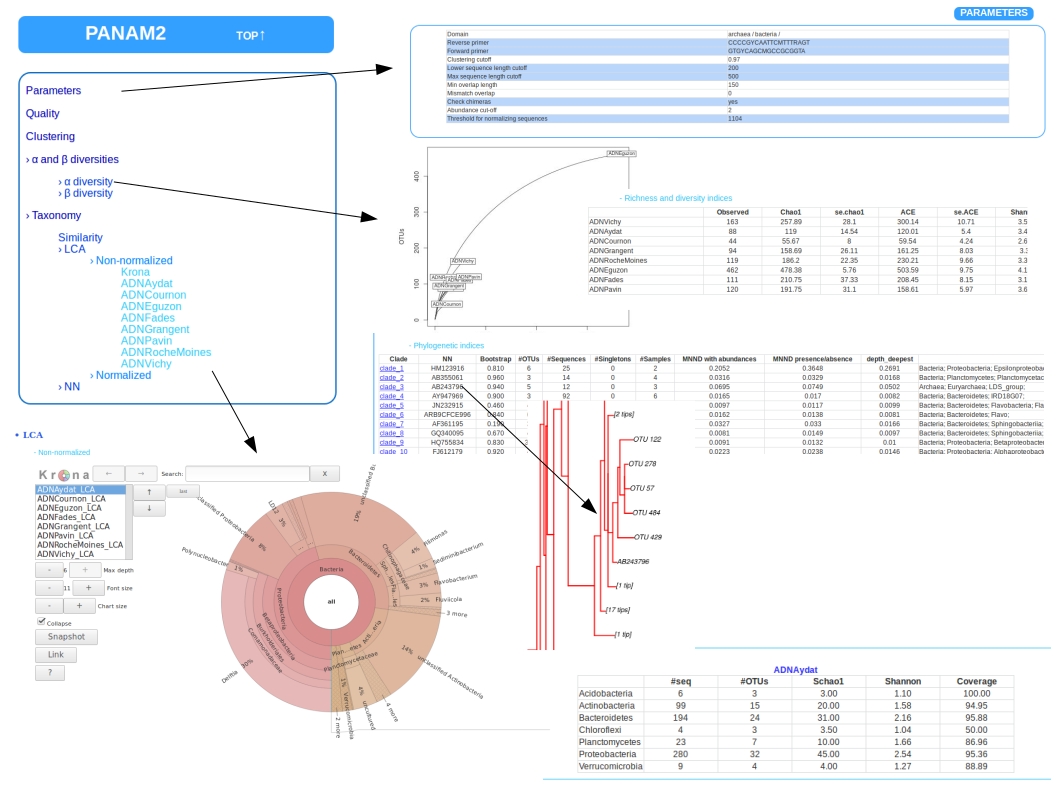
HTML REPORT
The HTML report allows to display the main results with a browser and can be open by typing in the result path:
Overview of the HTML report
INSTALLATION:
You can install PANAM2 in your Linux system (1) or a system "ready-to-use" withthis pipeline thanks to a docker image (2)
- download PANAM2 package in the final location folder and in a terminal type:
$ git clone https://github.com/panammeb/PANAM2.git # or download direcly from the web interface
$ cd PANAM2
$ perl setup.pl
VSEARCH, HMMALIGN, FASTTREE and Krona will be compiled and installed in the bin directory located in PANAM2. Perl and Bioperl are mandatory for the 3 main scripts listed above and have to be installed in the linux system with administrator privileges. R and packages Vegan, Phyloseq, Picante, Cluster and Mass must be also installed in your system for launching the script phylodiv_panam.pl and for processing some optionnal results generated by postprocess_panam.pl
All PANAM2 users must be able to write in the bd_ssrna directory: give the rights with the chmod command for multi-users access.
PANAM2 was tested on ubuntu system 16.10 with default packages.
- A docker image from this system with all dependances and the PANAM2 pipeline can be obtained from: https://www.dropbox.com/s/1byu667oota1skv/panam2.tar.bz2
Docker must be installed in your system
$ wget https://www.dropbox.com/s/1byu667oota1skv/panam2.tar.bz2
$ bzip2 -d panam2.tar.bz2
$ docker load < panam2.tar # the docker image loaded is named panam2:vXX with vXX the version number (v0.91 for example)
$ docker images # for checking the images loaded in the system
$ docker run -it panam2:vXX /bin/bash # this command allows to open the container and explore it with bash command but useless for using PANAM2 (see below)
USAGE:
- For the impatient
These different scripts can be launched by a unique command in a terminal with less oportunity for tuning analysis: panam2.pl
From the raw data included in the test directory (illumina sequencing of the V4 region of bacteria and archaea in lacustrine ecosystem)
This file can be with this command `wget https://www.dropbox.com/s/2y9p1jlfy00bsjq/test.tar.gz?dl=0`
$ perl panam2.pl -o example -r1 test/R1.fastq.gz -r2 test/R2.fastq.gz -f GTGYCAGCMGCCGCGGTA -r CCCCGYCAATTCMTTTRAGT -tags test/tags.txt -id 0.97 -dom bacteria -dom archaea -div Y # -div Y corresponds to the script phylodiv_panam.pl
From the raw data demultiplexed in the test2 directory (illumina sequencing of the V7 region of eukaryotes in lacustrin ecosystem)
The folder test2 consists of demultipled files with the following format <file name 1>_R1.fastq.gz <file name 1>_R2.fastq.gz / <file name 2>_R1.fastq.gz
$ perl panam2.pl -o example2 -d test2 -f GGCTTAATTTGACTCAACRCG -r GGGCATCACAGACCTGTTAT -id 0.95 -dom eukaryota
Help can be obtained by the following command:
$ perl panam2.pl -help
- Step by step
$ perl <name>_panam. pl file.ini
The file.ini includes all parameters describing the experiments (primers, tags, raw data...) and some methodological choices (threshold for clustering, parameters for merging, database used...).
Example: for the first and second step of the pipeline, type in a terminal:
$ perl quality_panam.pl test-paman.ini # this ini file is included in the package downloaded with all explanations
$ perl preprocess_panam.pl test-paman.ini
Results can be therefore analyzed step after step according to the options used (cleaning options for example). All the steps can be processed by the following command:
$ perl panam2.pl -ini file.ini
HTM report
$ firefox panam.html # Tested only with firefox
USAGE with the docker image:
From the user directory, type in a terminal:
$ docker run -v <user directory>:/share -it panam2:v0.94 perl PANAM2/panam2.pl -o /share/example2 -d share/test2 -f GGCTTAATTTGACTCAACRCG -r GGGCATCACAGACCTGTTAT -id 0.95 -dom eukaryota
-
:/share -> The user directory is shared in the "/share" path inside the container. For this example, the raw data are located in the path /test2 and the results will be generated in: /example2. In the container, these paths are: /share/test2 and /share/example2.
-
panam2:v0.91 -> image name (see installation procedure, this name can change according to the PANAM2 version)
-
perl PANAM2/panam2.pl -> perl scripts and all dependencies are installed in the PANAM2 directory in the container panam2:v0.91
MAIN FILES
HTML report is built from various tabulated files in the result directory
-
Clean demultiplexed sequences from the Illumina run: quality_output/seqAll.fasta
-
Sequences of representative OTUs processed from seqAll.fasta are in preprocess_output/pooled_sample/pooled_sample_SEQ_OTU. Tne seed OTUs are then assigned to their monophyletic taxonomic groups, aligned and a tree is built.
-
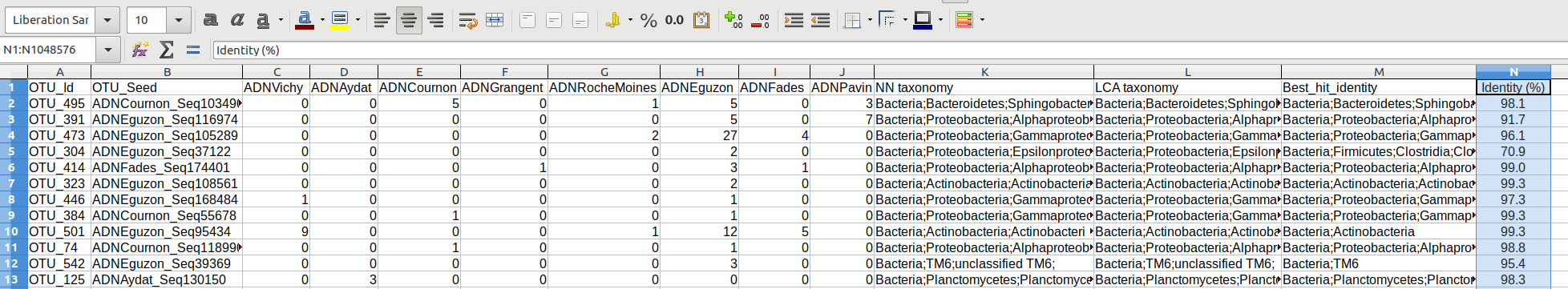
The main results are in the file named OTU_distribution_tax.txt: OTUs x samples x Taxonomies This tabulated file, located in the path results, can be open with any spreadsheet
-
OTU_distribution_tax_normalized_LCA.txt - OTU table normalized on the basis of the sample with the fewer number of reads (>1000) for the LCA taxonomy
-
OTU_distribution_tax_normalized_NN.txt - OTU table normalized on the basis of the sample with the fewer number of reads (>1000) for the LCA taxonomy
Richness and diversity indices can be found in the following files
-
taxonomic_distribution_LCA_norm.txt
-
taxonomic_distribution_LCA.txt
-
taxonomic_distribution_NN_norm.txt
-
taxonomic_distribution_NN.txt
CONTRIBUTORS:
- Didier Debroas (LMGE)
- Emilie Sicard (Internship Master Bioinformatics )
Based on: Taib N, Mangot J-F, Domaizon I, Bronner G, Debroas D. (2013). Phylogenetic Affiliation of SSU rRNA Genes Generated by Massively Parallel Sequencing: New Insights into the Freshwater Protist Diversity. PLoS ONE 8:e58950.