A short-read genotyper for various types of genetic variants (such as SNPs, indels and structural variants) represented in a pangenome graph. Genotypes are computed based on read k-mer counts and a panel of known haplotypes. A description of the method can be found here: https://doi.org/10.1038/s41588-022-01043-w
- conda or Singularity
Use the Singularity definition file located in container/ to build an (Ubuntu-based) container as follows (requires root privileges):
[sudo] singularity build pangenie.sif pangenie.def
In all usage examples below, call the PanGenie executable as follows:
singularity exec pangenie.sif PanGenie <PARAMETERS>
For example, to show PanGenie's command line help, use the following command:
singularity exec pangenie.sif PanGenie --help
You can check which versions of PanGenie (git hash) and of the jellyfish library have been installed in the container by running the following commands:
singularity exec pangenie.sif cat /metadata/jellyfish.lib.version
should produce a line like this (so, here, v2.3.0):
$ libjellyfish-2.0-2:amd64 2.3.0-4build1 libjellyfish-2.0-dev:amd64 2.3.0-4build1
singularity exec pangenie.sif cat /metadata/pangenie.git.version
should produce a line like this:
$ 5a1f9c5
git clone https://jana_ebler@bitbucket.org/jana_ebler/pangenie.git
cd pangenie
conda env create -f environment.yml
conda activate pangenie
mkdir build; cd build; cmake .. ; make
git clone https://jana_ebler@bitbucket.org/jana_ebler/pangenie.git
cd pangenie
mkdir build; cd build; cmake .. ; make
PanGenie is a pangenome-based genotyper using short-read data. It computes genotypes for variants represented as bubbles in a pangenome graph by taking information of already known haplotypes (represented as paths through the graph) into account. The required input files are described in detail below.
PanGenie expects a directed and acyclic pangenome graph as input (-v option).
This graph is represented in terms of a VCF file that needs to have certain properties:
- multi-sample - it needs to contain haplotype information of at least one known sample
- fully-phased - haplotype information of the known panel samples are represented by phased genotypes and each sample must be phased in a single block (i.e. from start to end).
- non-overlapping variants - the VCF represents a pangenome graph. Therefore, overlapping variation must be represented in a single, multi-allelic variant record.
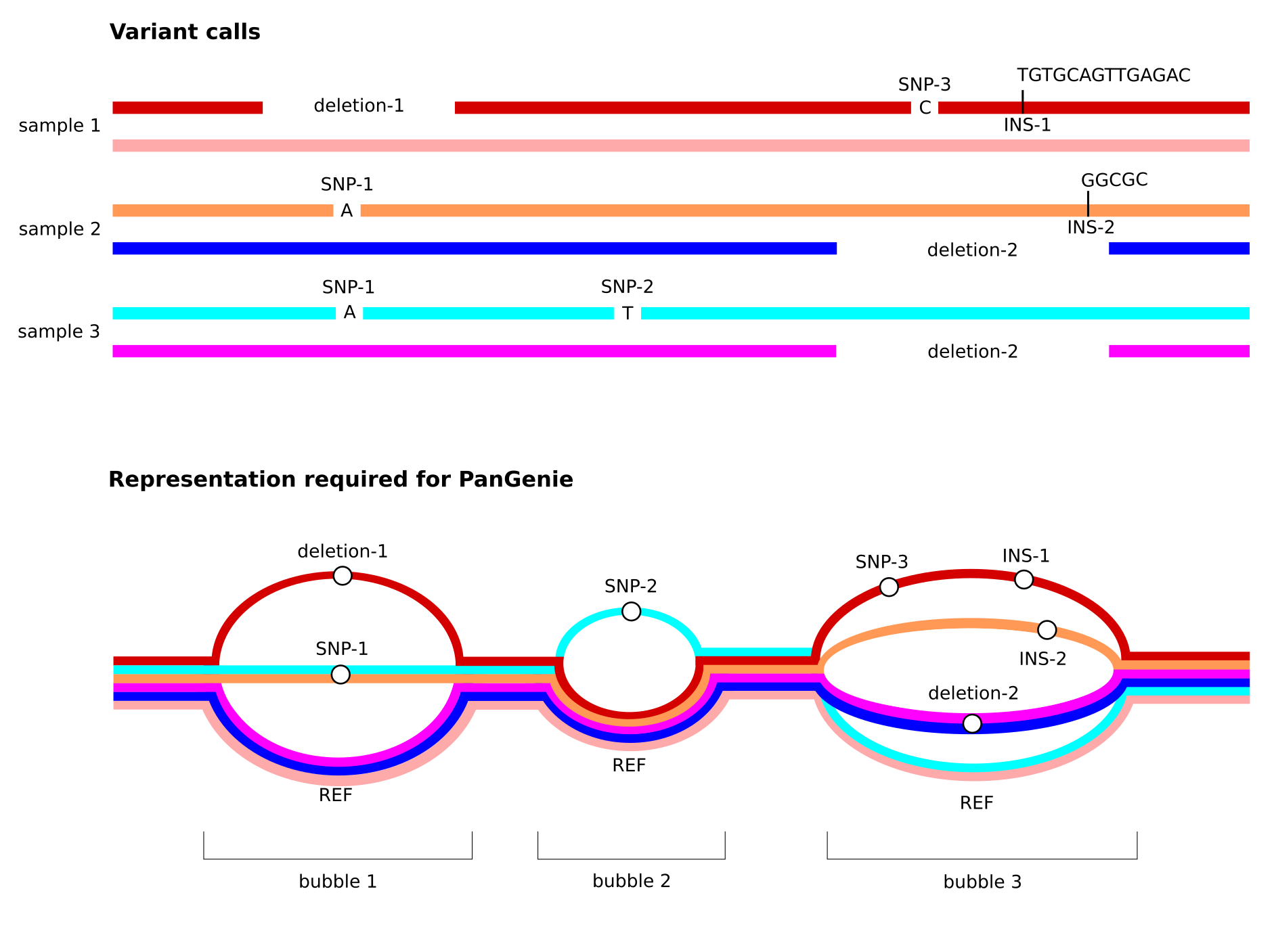
Note especially the third property listed above. See the figure below for an illustration of how overlapping variant alleles need to be represented in the input VCF provided to PanGenie.
We typically generate such VCFs from haplotype-resolved assemblies using this pipeline: https://bitbucket.org/jana_ebler/vcf-merging . However, any VCF with the properties listed above can be used as input.
In this case you can run PanGenie using the Snakemake pipeline provided in pipelines/run-from-callset/. This automatically merges overlapping alleles into mult-allelic VCF, runs PanGenie and later converts the output VCF back to the original representation.
PanGenie is k-mer based and thus expects short reads as input. Reads must be provided in a single FASTA or FASTQ file using the -i option.
PanGenie also needs a reference genome in FASTA format which can be provided using option -r.
PanGenie can be run using the command shown below:
./build/src/PanGenie -i <reads.fa/fq> -r <reference.fa> -v <variants.vcf> -t <nr threads for genotyping> -j <nr threads for k-mer counting>
The result will be a VCF file containing genotypes for the variants provided in the input VCF. Per default, the name of the output VCF is result_genotyping.vcf. You can specify the prefix of the output file using option -o <prefix>, i.e. the output file will be named as <prefix>_genotyping.vcf .
The full list of options is provided below.
program: PanGenie - genotyping and phasing based on kmer-counting and known haplotype sequences.
author: Jana Ebler
usage: PanGenie [options] -i <reads.fa/fq> -r <reference.fa> -v <variants.vcf>
options:
-c count all read kmers instead of only those located in graph.
-d do not add reference as additional path.
-e VAL size of hash used by jellyfish. (default: 3000000000).
-g run genotyping (Forward backward algorithm, default behaviour).
-i VAL sequencing reads in FASTA/FASTQ format or Jellyfish database in jf format.
NOTE: INPUT FASTA/Q FILE MUST NOT BE COMPRESSED. (required).
-j VAL number of threads to use for kmer-counting (default: 1).
-k VAL kmer size (default: 31).
-o VAL prefix of the output files. NOTE: the given path must not include non-existent folders. (default: result).
-p run phasing (Viterbi algorithm). Experimental feature.
-r VAL reference genome in FASTA format.
NOTE: INPUT FASTA FILE MUST NOT BE COMPRESSED. (required).
-s VAL name of the sample (will be used in the output VCFs) (default: sample).
-t VAL number of threads to use for core algorithm. Largest number of threads possible is the number of chromosomes given in the VCF (default: 1).
-u output genotype ./. for variants not covered by any unique kmers.
-v VAL variants in VCF format.
NOTE: INPUT VCF FILE MUST NOT BE COMPRESSED. (required).Runtime and memory usage depend on the number of variants genotyped and the number of haplotypes present in the graph.
With the data described here: https://doi.org/10.1038/s41588-022-01043-w, PanGenie ran in 1 hour and 25 minutes walltime using 22 cores (16 CPU hours) and used 68 GB RAM. The largest dataset that we have tested contained around 16M variants, 64 haplotypes and around 30x read coverage. Using 24 cores, PanGenie run in 1 hour and 46 minutes (24 CPU hours) and used 120 GB of RAM.
The largest panel we have run PanGenie on so far consisted of 44 samples (88 haplotypes). On this data, PanGenie needed 53 CPU hours (03:15 h wallclock time using 24 cores) and 153 GB of memory in order to genotype 20,661,169 variants.
The runtime of PanGenie gets slow as the number of haplotype paths increases. Due to technical reasons, the current implementation of PanGenie cannot handle more than 254 input haplotypes (127 diploid samples). In order to efficiently handle panels of this size and larger, the underlying model needs to be optimized.
The typical use case is to run PanGenie on a whole genome dataset. The following example is just a little demo illustrating how to run PanGenie.
We run PanGenie given a pangenome graph (VCF file,test-variants.vcf), sequencing reads (FASTA/FASTQ file, test-reads.fa) and a reference sequence (FASTA file, test-reference.fa) provided in the demo/ folder. After installation, PanGenie's genotyping algorithm can be run using the following command (which will take a few seconds for this example):
./build/src/PanGenie -i test-reads.fa -r test-reference.fa -v test-variants.vcf -o test -e 100000
The result will be a VCF file named test_genotyping.vcf containing the same variants as the input VCF with additional genotype predictions, genotype likelihoods and genotype qualities.
Parameter -e sets the hash size used by Jellyfish for k-mer counting. When running PanGenie on a whole genome dataset, this parameter can be omitted (so that PanGenie uses the default value).
Per default, PanGenie uses a single thread. The number of threads used for k-mer counting and genotyping/phasing can be set via parameters -j and -t, respectively.
J. Ebler, P. Ebert, W. E. Clarke, T. Rausch, P. A. Audano, T. Houwaart, Y. Mao, J. Korbel, E. E. Eichler, M. C. Zody, A. T. Dilthey, and T. Marschall. Pangenome-based genome inference. Nature genetics, 54(4):518–525, 2022.