DEVisComp
Description
DEVisComp is an R package for visualizing and comparing the
differential expression analysis results from R packages DESeq2 and
edgeR. This package provide different plots such as MA plots and Venn
diagrams comparing the differntially expressed genes data generated by
DESeq2 and edgeR. It can also be used to visualize the pattern of gene
expression together with log-fold change and adjusted p-values. In order
for the result to be meaningful, the package assumes the results from
DESeq2 and edgeR should be created with similar parameters.
The package is developed under R 4.1.1 in Mac.
Installation
To install the latest version of the package:
require("devtools")
devtools::install_github("Lori-tan/DEVisComp", build_vignettes = TRUE)
library("DEVisComp")To run the Shiny app:
runDEVisComp()Overview
ls("package:DEVisComp")
data(package = "DEVisComp") # optionalDEVisComp contains 1 functions to visualize the expression analysis
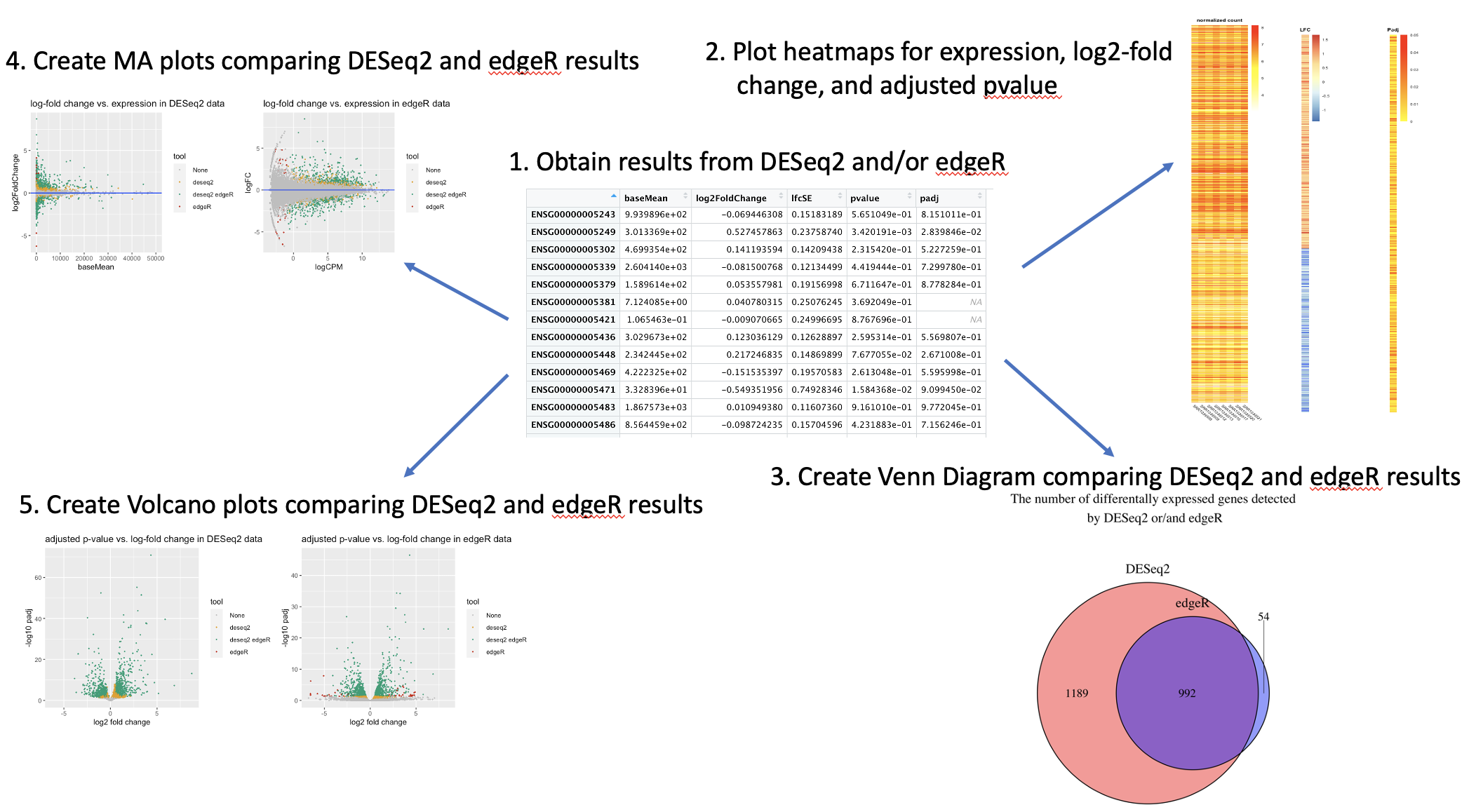
results and 3 functions to compare the results created by DESeq2 and
edgeR. The ClusterTogether function generates 3 heatmaps side-by-side
showing the patterns among gene expression, log-fold changes, and
adjusted p-values of the differntially expressed genes. The compVenn
is the function that create a Venn diagram comparing the genes that are
marked as differentially expressed by DESeq2 and edgeR. The compMA and
compVolcano generate MA plots and Volcano plots respectively for
results created by DESeq2 and edgeR.
browseVignettes("DEVisComp")An overview of the package is illustrated below.
Contributions
The author of the package is Luomeng Tan. The ClusterTogether function
makes use of pheatmap function from pheatmap R package to plot the
heatmaps. The stats R package is used for calculating the standard
deviation between counts in different condition and for calculating the
quantile of a vector. The RColorBrewer and grDevices R packages are
used to generate color scale for the heatmaps. The cowplot R packages
is used to plot mulitple heatmaps into a grid. The compVenn makes use
of the VennDiagram R package. compMA and compVolcano use ggplot2
and use gridExtra to plot plot multiple plots into one grid. The
package uses data from an RNAseq experiment characterize the human
airway smooth muscle transcriptome conducted by University of
Pennsylvania as examples and exports them as data available to users.
References
Auguie, B. (2017). gridExtra: Miscellaneous Functions for “Grid” Graphics. R package version 2.3. https://CRAN.R-project.org/package=gridExtra
Chen, H. (2021). VennDiagram: Generate High-Resolution Venn and Euler Plots. R package version 1.7.0. https://CRAN.R-project.org/package=VennDiagram
Himes et al. (2014). RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PloS one, 9(6), e99625. https://doi.org/10.1371/journal.pone.0099625
Kolde, R. (2019). pheatmap: Pretty Heatmaps. R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap
Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 550. https://doi.org/10.1186/s13059-014-0550-8
Neuwirth, E. (2014). RColorBrewer: ColorBrewer Palettes. R package version 1.1-2. https://CRAN.R-project.org/package=RColorBrewer
R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Robinson, M. D., McCarthy, D. J., & Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. https://doi.org/10.1093/bioinformatics/btp616
Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York.
Wilke, C. O. (2020). cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’. R package version 1.1.1. https://CRAN.R-project.org/package=cowplot
Zhu, A., Ibrahim, J. G., Love, M. I. (2018). Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences Bioinformatics
Acknowledgements
This package was developed as part of an assessment for 2021 BCB410H: Applied Bioinfor- matics, University of Toronto, Toronto, CANADA.