Scripts used for computing Flux of Genes Segments (FOGS), which enable to quantify the rate of gene acquisition or loss events within a collection of bacterial genomes.
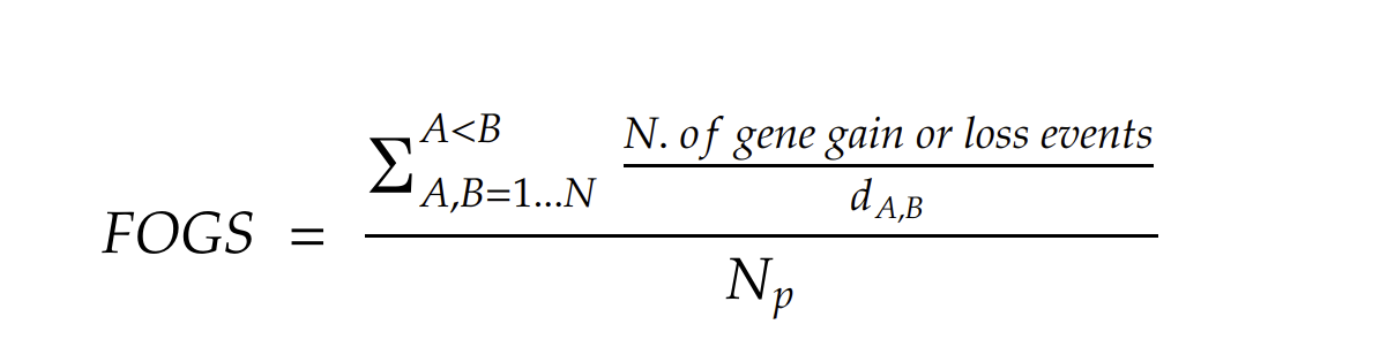 d is the evolutionary distance between genome A and genome B, Np is the total number of genome pairs considered (Np = 2/[N(N-1)], where N is the number of genomes considered). The higher the value of FOGS, the higher is the genome plasticity.
d is the evolutionary distance between genome A and genome B, Np is the total number of genome pairs considered (Np = 2/[N(N-1)], where N is the number of genomes considered). The higher the value of FOGS, the higher is the genome plasticity.
- Linux or macOS
- Python 3.1 or later
- Python libraries: pandas, scipy, multiprocessing, seaborn, matplotlib
- Inputs required for computing the number of gene gain and loss events are:
- A GFF file for each genome, both prodigal and prokka output are accepted
- Orthology groups, formatted as follow [orthology_groups_file]:
orthologygroup1: GFFID_1 GFFID_112 GFFID_2 GFFID_90 GFFID_1872
orthologygroup2: GFFID_2 GFFID_1989 GFFID_2131 GFFID_1213 GFFID_5090
- A tsv table containing the evolutionary distance for each genomes combination [evolutionary_distaces.tsv]:
genomeA genomeB 12
genomeA genomeC 23
If you are considering closely related strains (e.g. same ST), we suggest to use SNP distances. In the original work, P-DOR was used to obtain the SNP alignment, and snp-dist (-m) to obtain the tsv file.
- A tsv table containing the sub-partitioning for the dataset [clusters.tsv]:
genome Clone
genomeA cluster1
genomeB cluster2
- Compute the number of gene gain and loss events for each combination of genomes. All the inputs (gff files and orthology groups file) and the script should be in the same folder
python3 gainlossevents.py orthology_groups_fileA csv files (GainLossEvents.csv) containing the number of gene gain/loss events of each genome is produced.
- The number of gene gain or loss events is then weighted on the SNP distance and FOGS value is produced for each cluster
python3 compute_FOGS.py clusters.tsv evolutionary_distaces.tsv
FOGS.tsv contains the FOGS value for each cluster in the dataset