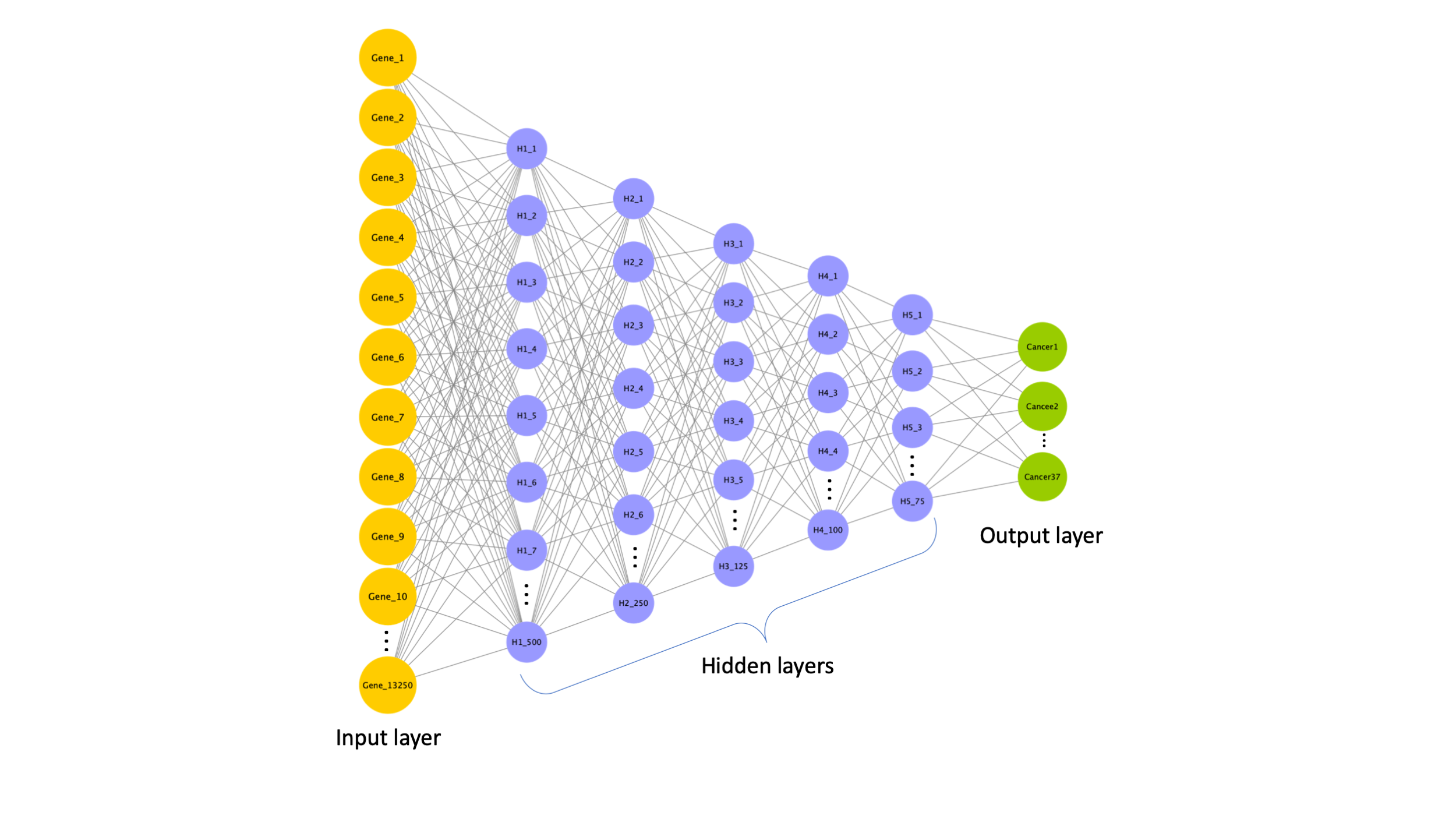
This repository contains code used to build and interpret a deep learning model. It is a DNN classifier trained using gene expression data (TCGA). Then is interpreted to identify cancer specific gene expression signatures.
repository contains following jupyter notebooksi and should be used in order below,
- Model.ipynb
- train test deep learning models
- ShapInterpretation.ipynb
- shap value calculation for each feature
- MergeShapChunks.ipynb
- merge shap files to create one file per model
- ShapTopGenes.ipynb
- Get top 20 genes for each cancer types
- SelectGeneSignaturesFC.ipynb
- Filtering of gene to produce final list of gene signatures