Quantiles Regression is Poorly-Calibrated
joseortiz3 opened this issue · 10 comments
Environment info
Operating System: Windows 10 64
CPU: i7-7700k
C++/Python/R version: Python 3.6 Anaconda 5.0.0
Problem Description
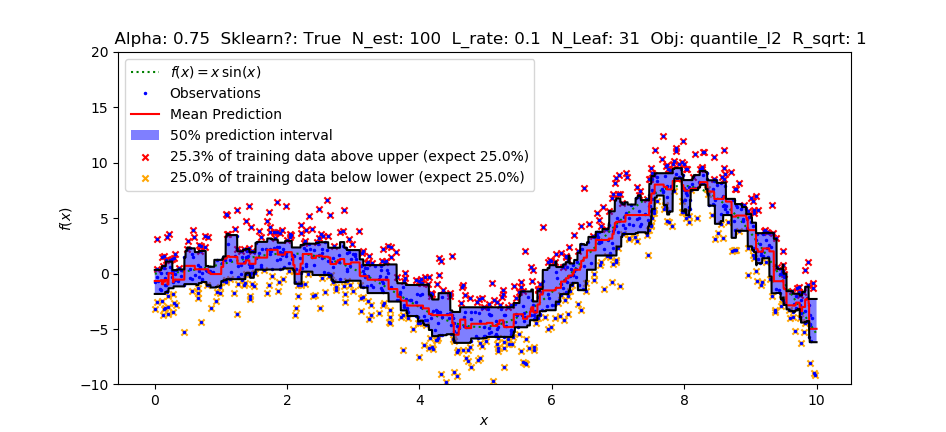
The quantile-estimation functionality recently implemented is poorly-calibrated in comparison to sklearn's GradientBoostingRegressor. This means that specifying the quantile (75% percentile/quantile, for instance) results in estimations that do not bound 75% of the training data (usually less in practice), and no configuration fixes this.
Reproducible examples
I have modified the script given in a previous issue on quantiles. Toggle USE_SKLEARN to observe issue. Change parameters to see that calibration problem persists.
import numpy as np
import matplotlib.pyplot as plt
import lightgbm as lgb
from sklearn.ensemble import GradientBoostingRegressor
from gbdt_quantiles import plot_figure
np.random.seed(1)
# Use sklearn or lightgbm?
USE_SKLEARN = True # Toggle this to observe issue.
# Quantile to Estimate
alpha = 0.75
# Training data size
N_DATA = 1000
# Function to Estimate
def f(x):
"""The function to predict."""
return x * np.sin(x)
# model parameters
LEARNING_RATE = 0.1
N_ESTIMATORS = 100
MAX_DEPTH = -1
NUM_LEAVES = 31 # lgbm only
OBJECTIVE = 'quantile_l2' # lgbm only, 'quantile' or 'quantile_l2'
REG_SQRT = True # lgbm only
if USE_SKLEARN:
if MAX_DEPTH < 0: # sklearn grows differently than lgbm.
print('Max Depth specified is incompatible with sklearn. Changing to 3.')
MAX_DEPTH = 3
#---------------------- DATA GENERATION ------------------- #
# First the noiseless case
X = np.atleast_2d(np.random.uniform(0, 10.0, size=N_DATA)).T
X = X.astype(np.float32)
# Observations
y = f(X).ravel()
dy = 1.5 + 1.0 * np.random.random(y.shape)
noise = np.random.normal(0, dy)
y += noise
y = y.astype(np.float32)
# Mesh the input space for evaluations of the real function, the prediction and
# its MSE
xx = np.atleast_2d(np.linspace(0, 10, 9999)).T
xx = xx.astype(np.float32)
# Train high, low, and mean regressors.
# ------------------- HIGH/UPPER BOUND ------------------- #
if USE_SKLEARN:
clfh = GradientBoostingRegressor(loss='quantile', alpha=alpha,
n_estimators=N_ESTIMATORS, max_depth=MAX_DEPTH,
learning_rate=LEARNING_RATE, min_samples_leaf=9,
min_samples_split=9)
clfh.fit(X, y)
else:
## ADDED
clfh = lgb.LGBMRegressor(objective = OBJECTIVE,
alpha = alpha,
num_leaves = NUM_LEAVES,
learning_rate = LEARNING_RATE,
n_estimators = N_ESTIMATORS,
reg_sqrt = REG_SQRT,
max_depth = MAX_DEPTH)
clfh.fit(X, y,
#eval_set=[(X, y)],
#eval_metric='quantile'
)
## END ADDED
# ------------------- LOW/LOWER BOUND ------------------- #
if USE_SKLEARN:
clfl = GradientBoostingRegressor(loss='quantile', alpha=1.0-alpha,
n_estimators=N_ESTIMATORS, max_depth=MAX_DEPTH,
learning_rate=LEARNING_RATE, min_samples_leaf=9,
min_samples_split=9)
clfl.fit(X, y)
else:
## ADDED
clfl = lgb.LGBMRegressor(objective = OBJECTIVE,
alpha = 1.0 - alpha,
num_leaves = NUM_LEAVES,
learning_rate = LEARNING_RATE,
n_estimators = N_ESTIMATORS,
reg_sqrt = REG_SQRT,
max_depth = MAX_DEPTH)
clfl.fit(X, y,
#eval_set=[(X, y)],
#eval_metric='quantile'
)
## END ADDED
# ------------------- MEAN/PREDICTION ------------------- #
if USE_SKLEARN:
clf = GradientBoostingRegressor(loss='ls',
n_estimators=N_ESTIMATORS, max_depth=MAX_DEPTH,
learning_rate=LEARNING_RATE, min_samples_leaf=9,
min_samples_split=9)
clf.fit(X, y)
else:
## ADDED
clf = lgb.LGBMRegressor(objective = 'regression',
num_leaves = NUM_LEAVES,
learning_rate = LEARNING_RATE,
n_estimators = N_ESTIMATORS,
max_depth = MAX_DEPTH)
clf.fit(X, y,
#eval_set=[(X, y)],
#eval_metric='l2',
#early_stopping_rounds=5
)
## END ADDED
# ---------------- PREDICTING ----------------- #
# Make the prediction on the meshed x-axis
y_pred = clf.predict(xx)
y_lower = clfl.predict(xx)
y_upper = clfh.predict(xx)
# Check calibration by predicting the training data.
y_autopred = clf.predict(X)
y_autolow = clfl.predict(X)
y_autohigh = clfh.predict(X)
frac_below_upper = round(np.count_nonzero(y_autohigh > y) / len(y),3)
frac_above_upper = round(np.count_nonzero(y_autohigh < y) / len(y),3)
frac_above_lower = round(np.count_nonzero(y_autolow < y) / len(y),3)
frac_below_lower = round(np.count_nonzero(y_autolow > y) / len(y),3)
# Print calibration test
print('fraction below upper estimate: \t actual: ' + str(frac_below_upper) + '\t ideal: ' + str(alpha))
print('fraction above lower estimate: \t actual: ' + str(frac_above_lower) + '\t ideal: ' + str(alpha))
# ------------------- PLOTTING ----------------- #
plt.plot(xx, f(xx), 'g:', label=u'$f(x) = x\,\sin(x)$')
plt.plot(X, y, 'b.', markersize=3, label=u'Observations')
plt.plot(xx, y_pred, 'r-', label=u'Mean Prediction')
plt.plot(xx, y_upper, 'k-')
plt.plot(xx, y_lower, 'k-')
plt.fill(np.concatenate([xx, xx[::-1]]),
np.concatenate([y_upper, y_lower[::-1]]),
alpha=.5, fc='b', ec='None', label=(str(round(100*(alpha-0.5)*2))+'% prediction interval'))
plt.scatter(x=X[y_autohigh < y], y=y[y_autohigh < y], s=20, marker='x', c = 'red',
label = str(round(100*frac_above_upper,1))+'% of training data above upper (expect '+str(round(100*(1-alpha),1))+'%)')
plt.scatter(x=X[y_autolow > y], y=y[y_autolow > y], s=20, marker='x', c = 'orange',
label = str(round(100*frac_below_lower,1))+ '% of training data below lower (expect '+str(round(100*(1-alpha),1))+'%)')
plt.xlabel('$x$')
plt.ylabel('$f(x)$')
plt.ylim(-10, 20)
plt.legend(loc='upper left')
plt.title( ' Alpha: '+str(alpha) +
' Sklearn?: '+str(USE_SKLEARN) +
' N_est: '+str(N_ESTIMATORS) +
' L_rate: '+str(LEARNING_RATE) +
' N_Leaf: '+str(NUM_LEAVES) +
' Obj: '+str(OBJECTIVE) +
' R_sqrt: '+str(int(REG_SQRT))
)
plt.show()Steps to reproduce
- Run code snippet with USE_SKLEARN set to True
- Run again with USE_SKLEARN set to False
- Compare Results
- Modify parameters, repeat, etc.
Results
As you can see above, LightGBM's implementation of quantiles is estimating a narrower quantile (about .62) than was specified (.75). Sklearn on the other hand produces a well-calibrated quantile estimate. Playing with the parameters does not help.
@joseortiz3
Thanks, this is very helpful. It indeed is a issue.
As we discussed in #1109 (comment) , current implementation of quantile in LightGBM is not perfect.
There are 2 reasons:
- the gradient/hessian of quantile loss is not easy to fit.
quantile_l2is a trade-off solution, which is not equivalent toquantile. Maybe you can try this: (https://www.bigdatarepublic.nl/regression-prediction-intervals-with-xgboost/) and provides some feedback. - The leaf-value in LightGBM is sum_grad/sum_hess, while it is the value of corresponding quantile in sklearn. Sklearn's solution seems better. Maybe we can try something similar.
@Laurae2 @henry0312
any suggestions for the quantile objective ?
"Maybe you can try this: (https://www.bigdatarepublic.nl/regression-prediction-intervals-with-xgboost/) and provides some feedback."
Oh, by the way, yes I've seen this. That approach is not very useful for me (finance, science). Grid searching three parameters to find a well-calibrated quantile estimate is wasteful, and sklearn's implementation proves that a well-calibrated quantile estimate is possible without such a search. Sklearn's estimation is remarkably robust when estimating vastly different things / different distributions, so whatever they are doing is the way to go judging from my experience.
@joseortiz3 Thanks for feedback.
So maybe we should try to fix the leaf-value towards sklearn's solution.
@guolinke The user must transform the labels to [0, 1] for quantile regression, then extrapolate back to the original range of the labels.
Quantile regression in LightGBM will not work properly without scaling values to the correct range. For instance, scikit-learn uses the range [0, 100], with alpha*100 = alpha for the target quantile. Training is not done using the labels, but is done using the labels' quantiles.
@Laurae2 I didn't find where sklearn normalize the range of label to [0,100].
Can you provide some sources of it ?
@guolinke It does quantile transformation here: https://github.com/scikit-learn/scikit-learn/blob/master/sklearn/ensemble/gradient_boosting.py#L434-L442
@Laurae2
I think it is not the same as normalizing label range to [0,100].
sklearn assigns the percentile value of delta(y-pred_over_previous_trees) for each leaf.
I think simply normalize the label range range cannot help, since the leaf output is still the sum_grad/sum_hess, which is not same as the percentile value even for normalized label.
I think now with the new updates that fix this issue, LightGBM is the fastest, quantiles-supporting boosted decision tree implementation available. Pretty exciting! I'm getting about 20x speedup with similar performance over sklearn in quantile workloads! Great work.