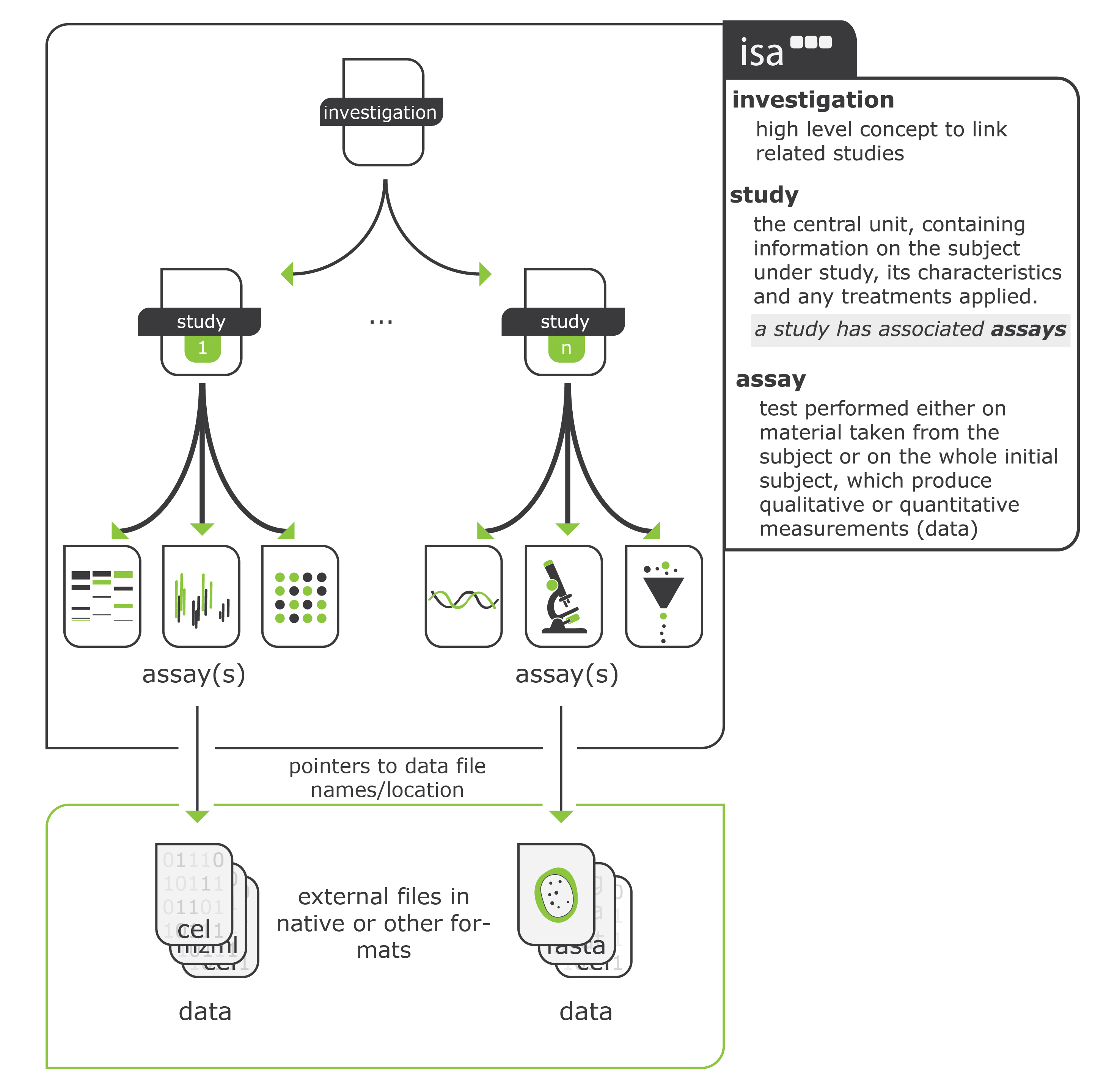
linked from https://isa-tools.org/format/specification.html
pISA-tree: Standard project directory tree (ISA-tab compliant)
https://github.com/NIB-SI/pISA
pISA-tree: Standard project directory tree (ISA-tab compliant)
https://github.com/NIB-SI/seekr
R support for pISA-tree
https://github.com/NIB-SI/pisar
FAIRDOMHub: a repository and collaboration environment for sharing systems biology research
LTS versions of:
apt-key adv --keyserver [from this location or server] --recv-keys [retrieve key(s)]
add-apt-repository ‘deb https://cloud.r-project.org/bin/linux/ubuntu [type appropriate selection from https://cloud.r-project.org/bin/linux/ubuntu/]’
apt-get update
apt-get install r-base
apt-get install r-base-dev
apt install build-essential
apt-get install pandoc
R
q()
R --help
R -e 'install.packages("rmarkdown", repos="https://cran.rstudio.com/")'
R -e "shiny::runApp('./pathToShinyApp/name.R')"
Rscript -e "rmarkdown::render('./pathToScript/scriptName.Rmd')"
pip install biopython
apt-get install dos2unix
apt-get install moreutils
R -e 'install.packages(c("Rcpp", "httpuv"))'
R -e 'install.packages("shiny")'
R -e 'install.packages("devtools", repos="https://cran.rstudio.com/")'
apt-get install libssl-dev libxml2-dev libcurl4-openssl-dev libcurl4-gnutls-dev curl
linked from www.git-tower.com
always use relative path
. represents the current directory
.. represents the parent directory
ls .. list information about the files one level up,
ls ../.. or ls ../../ list information about the files two levels up, etc
- use wget to pull down the
rawfile
$ wget https://raw.githubusercontent.com/username/reponame/path/to/file
- use git clone to pull the complete repository (prerequisites: user with
sudoprivileges)
$ sudo apt update
$ sudo apt install git
$ git clone https://github.com/username/reponame.git
- Install Perl on Ubuntu-like Linux OS (prerequisites: user with
sudoprivileges)
$ sudo apt update
$ sudo apt-get install perl
# check version
$ perl -v
More details for installing Perl can be found at perl.org
-
Use CPAN (‘Comprehensive Perl Archive Network’) to install Perl modules
-
Install different version of Perl
$ sudo cpan App::perlbrew
$ perlbrew init
# see which versions are available:
$ perlbrew available
# install version 5.X.Y
$ perlbrew install perl-5.X.Y
# list all installed versions
$ perlbrew list
# change Perl for the current shell # (or per your sessions)
$ perlbrew use perl-5.X.Y # (or perlbrew switch perl-5.X.Y)
$ which perl
# revert version to default for the current shell # (or per your sessions)
$ perlbrew off # (or perlbrew switch-off)
Installing pip for Python 3 and Python2 on Ubuntu-like OS (prerequisites: user with sudo privileges)
# Installing pip for Python 3
$ sudo apt update
$ sudo apt install python3-pip
# Installing pip for Python 2
$ sudo apt update
$ sudo apt install python2
$ curl https://bootstrap.pypa.io/get-pip.py --output get-pip.py
$ sudo python2 get-pip.py