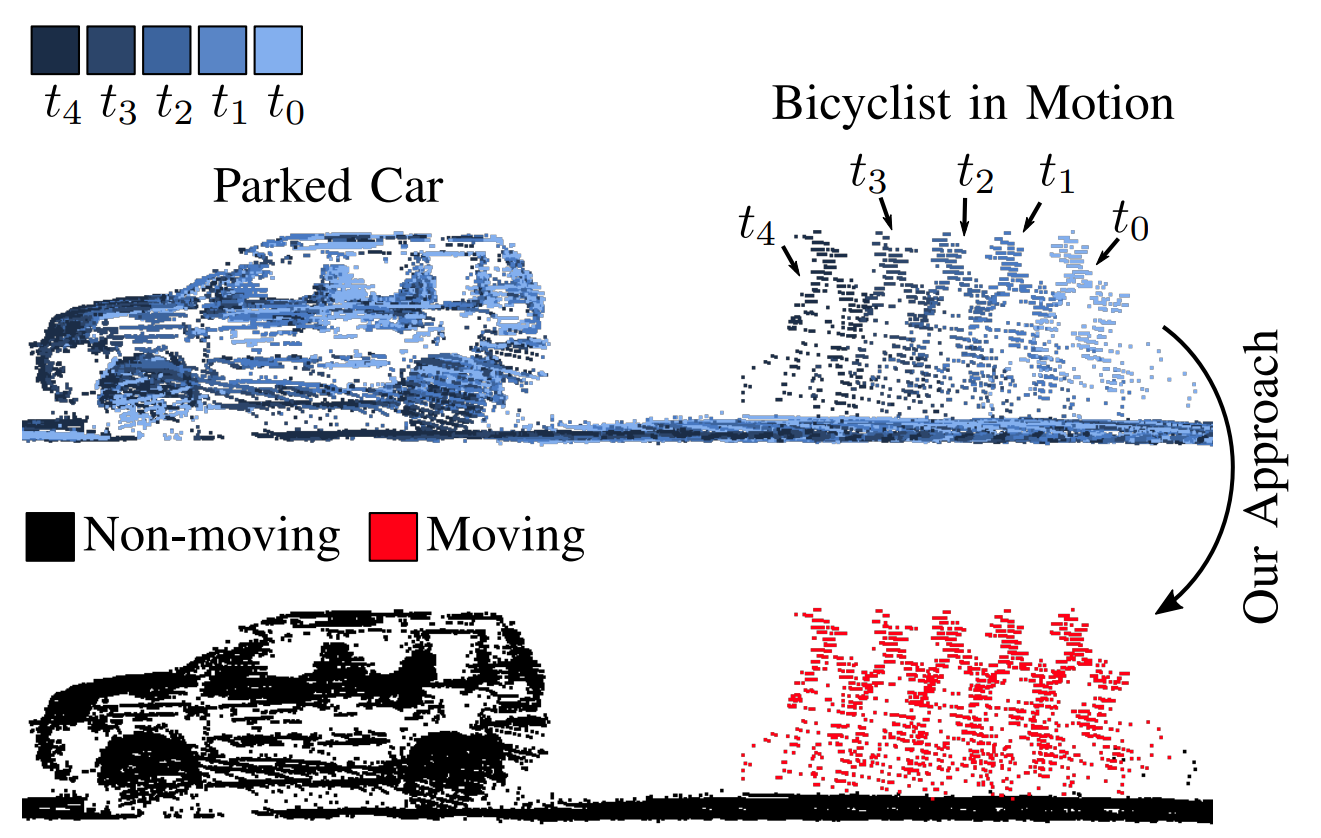
Our moving object segmentation on the unseen SemanticKITTI test sequences 18 and 21. Red points are predicted as moving.
Our moving object segmentation on the unseen SemanticKITTI test sequences 18 and 21. Red points are predicted as moving.
- Publication
- Overview
- Data
- Installation
- Running the Code
- Evaluation and Visualization
- Benchmark
- Pretrained Model
- License
If you use our code in your academic work, please cite the corresponding paper:
@article{mersch2022ral,
author = {B. Mersch and X. Chen and I. Vizzo and L. Nunes and J. Behley and C. Stachniss},
title = {{Receding Moving Object Segmentation in 3D LiDAR Data Using Sparse 4D Convolutions}},
journal={IEEE Robotics and Automation Letters (RA-L)},
year = 2022,
volume = {7},
number = {3},
pages = {7503--7510},
}Please find the corresponding video here.
Given a sequence of point clouds, our method segments moving (red) from non-moving (black) points.
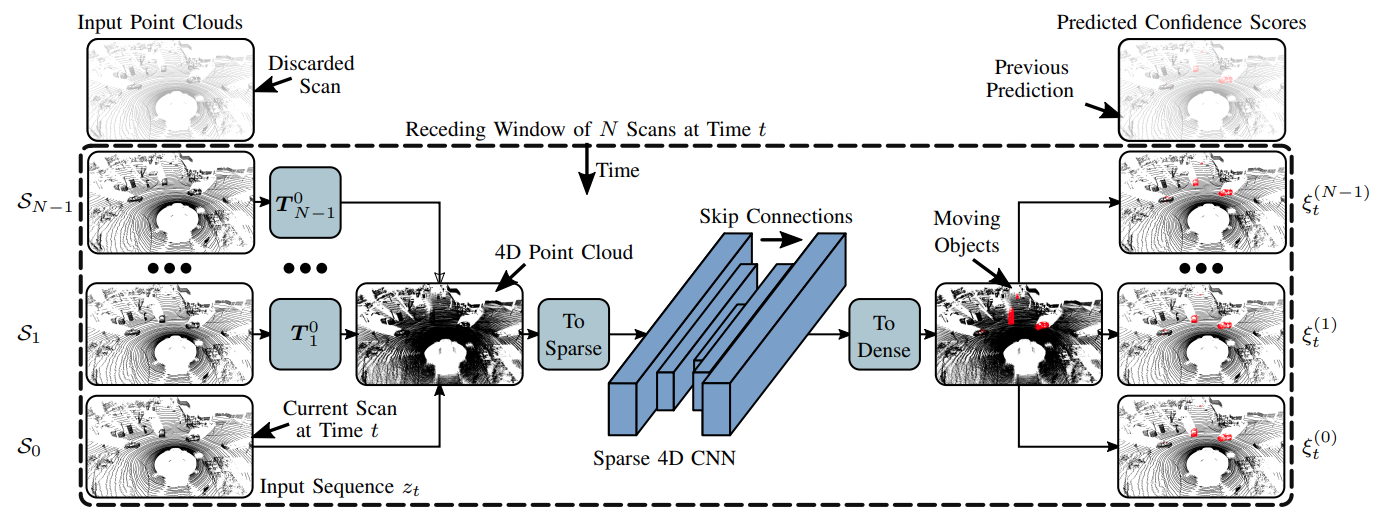
We first create a sparse 4D point cloud of all points in a given receding window. We use sparse 4D convolutions from the MinkowskiEngine to extract spatio-temporal features and predict per-points moving object scores.
Download the SemanticKITTI data from the official website.
./
└── sequences
├── 00/
│ ├── velodyne/
| | ├── 000000.bin
| | ├── 000001.bin
| | └── ...
│ └── labels/
| ├── 000000.label
| ├── 000001.label
| └── ...
├── 01/ # 00-10 for training
├── 08/ # for validation
├── 11/ # 11-21 for testing
└── ...
Clone this repository in your workspace with
git clone https://github.com/PRBonn/4DMOS
We provide a Dockerfile and a docker-compose.yaml to run all docker commands with a simple Makefile.
To use it, you need to
-
In Ubuntu, install docker-compose with
sudo apt-get install docker-compose
Note that this will install docker-compose v1.25 which is recommended since GPU access during build time using docker-compose v2 is currently an open issue.
-
Install the NVIDIA Container Toolkit
-
IMPORTANT To have GPU access during the build stage, make
nvidiathe default runtime in/etc/docker/daemon.json:{ "runtimes": { "nvidia": { "path": "/usr/bin/nvidia-container-runtime", "runtimeArgs": [] } }, "default-runtime": "nvidia" }
Save the file and run
sudo systemctl restart dockerto restart docker. -
Build the image with all dependendencies with
make build
Before running the container, you need to set the path to your dataset:
export DATA=path/to/dataset/sequencesTo test that your container is running propoerly, do
make testFinally, run the container with
make runYou can now work inside the container and run the training and inference scripts.
Without Docker, you need to install the dependencies specified in the setup.py. This can be done in editable mode by running
python3 -m pip install --editable .Now install the MinkowskiEngine according to their installation wiki page. When installing the MinkowskiEngine, your CUDA version has to match the CUDA version that was used to compile PyTorch.
If not done yet, specify the path to the SemanticKITTI data:
export DATA=path/to/dataset/sequencesIf you use Docker, you now need to run the container with make run.
To train a model with the parameters specified in config/config.yaml, run
python scripts/train.pyFind more options like loading weights from a pre-trained model or checkpointing by passing the --help flag to the command above.
Inference is done in two steps. First, predicting moving object confidence scores and second, fusing multiple confidence values to get a final prediction (non-overlapping strategy or binary Bayes filter.
To infer the per-point confidence scores for a model checkpoint at path/to/model.ckpt, run
python scripts/predict_confidences.py -w path/to/model.ckptWe provide several additional options, see --help flag. The confidence scores are stored in predictions/ID/POSES/confidences to distinguish setups using different model IDs and pose files.
Next, the final moving object predictions can be obtained by
python scripts/confidences_to_labels.py -p predictions/ID/POSESYou can use the --strategy argument to decide between the non-overlapping or bayesian filter strategy from the paper. Run with --help to see more options. The final predictions are stored in predictions/ID/POSES/labels/.
We use the SemanticKITTI API to evaluate the intersection-over-union (IOU) of the moving class as well as to visualize the predictions. Clone the repository in your workspace, install the dependencies and then run the following command to visualize your predictions for e.g. sequence 8:
cd semantic-kitti-api
./visualize_mos.py --sequence 8 --dataset /path/to/dataset --predictions /path/to/4DMOS/predictions/ID/POSES/labels/STRATEGY/
To submit the results to the LiDAR-MOS benchmark, please follow the instructions here.
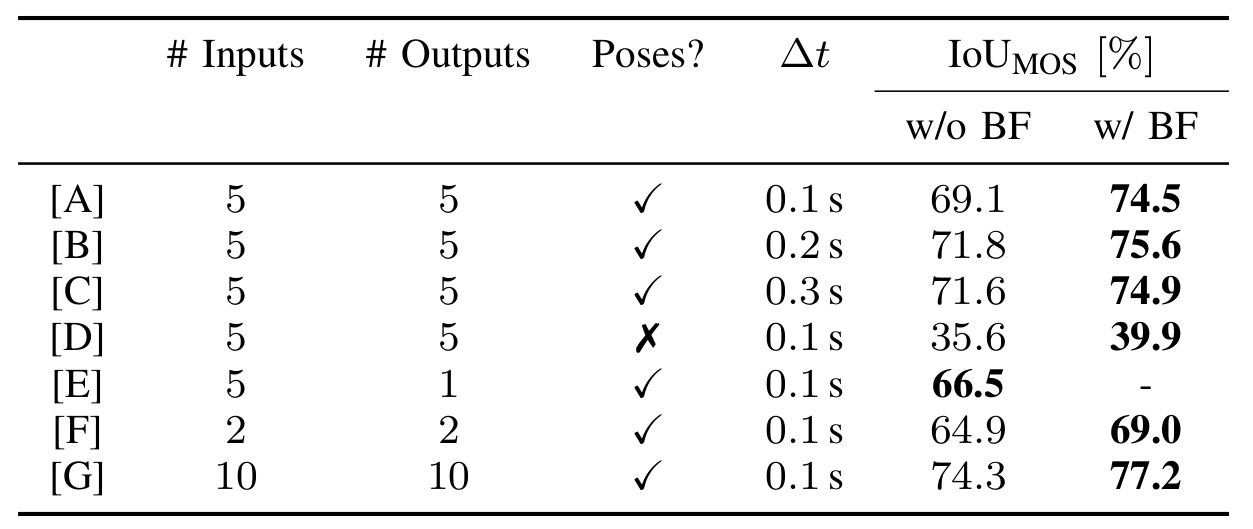
- Model [A]: 5 scans @ 0.1s
- Model [B]: 5 scans @ 0.2s
- Model [C]: 5 scans @ 0.3s
- Model [D]: 5 scans, no poses
- Model [E]: 5 scans input, 1 scan output
- Model [F]: 2 scans
- Model [G]: 10 scans
This project is free software made available under the MIT License. For details see the LICENSE file.