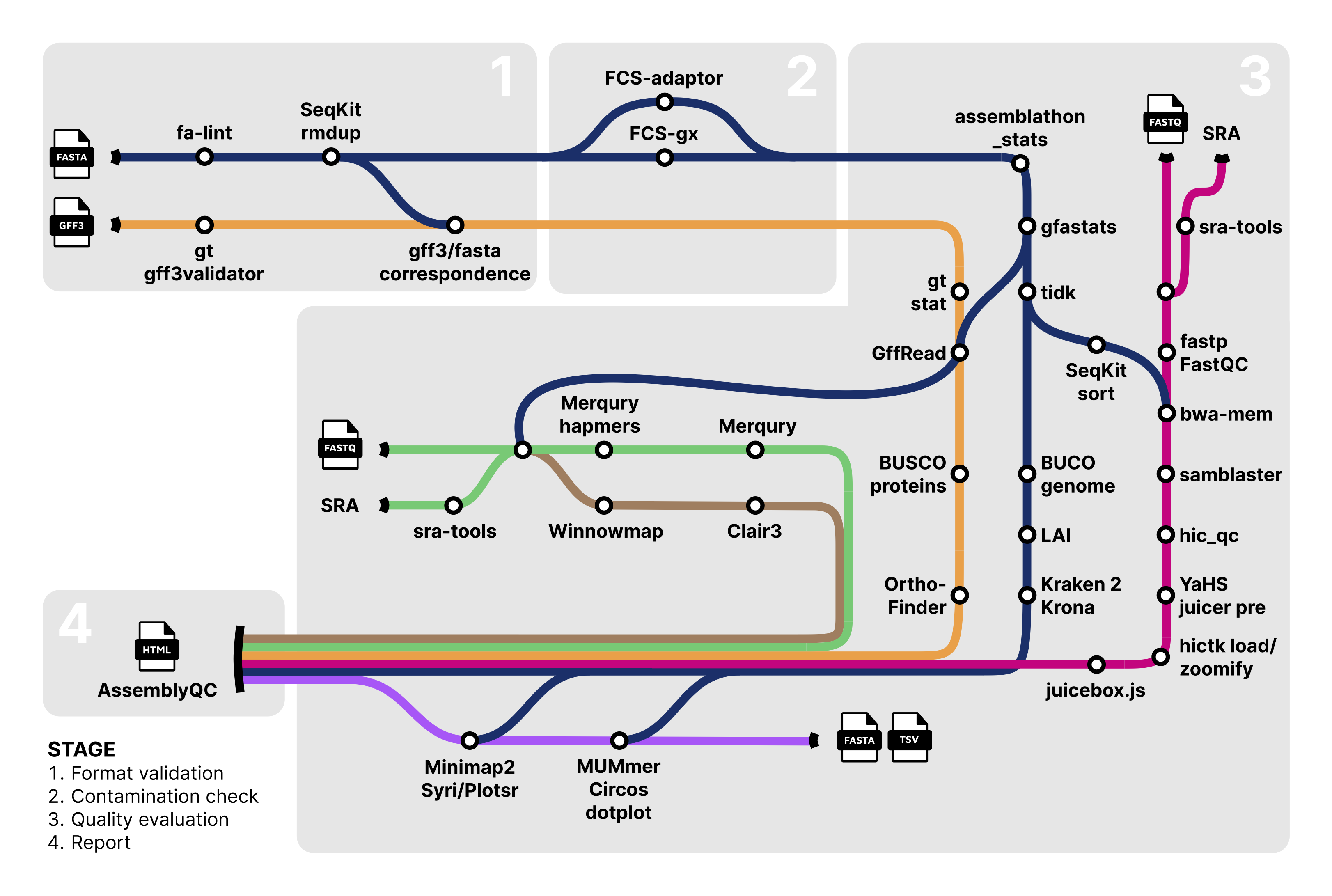
plant-food-research-open/assemblyqc is a Nextflow pipeline which evaluates assembly quality with multiple QC tools and presents the results in a unified html report. The tools are shown in the Pipeline Flowchart and their references are listed in CITATIONS.md. The pipeline includes skip flags to disable execution of various tools.
Assembly- fasta_validator + SeqKit rmdup: FASTA validation
- assemblathon_stats, gfastats: Assembly statistics
- NCBI FCS-adaptor: Adaptor contamination pass/fail
- NCBI FCS-GX: Foreign organism contamination pass/fail
- tidk: Telomere repeat identification
- BUSCO: Gene-space completeness estimation
- LAI: Continuity of repetitive sequences
- Kraken 2, Krona: Taxonomy classification
Alignment and visualisation of HiC data- sra-tools: HiC data download from SRA or use of local FASTQ files
- fastp, FastQC: Read QC and trimming
- SeqKit sort: Alphanumeric sorting of FASTA by sequence ID
- bwa-mem: HiC read alignment
- samblaster: Duplicate marking
- hic_qc: HiC read and alignment statistics
- Matlock: BAM to juicer conversion
- 3d-dna/visualize:
.hicfile creation - juicebox.js: HiC contact map visualisation
K-mer completeness, consensus quality and phasing assessment- sra-tools: Assembly, maternal and paternal data download from SRA or use of local FASTQ files
- Merqury hapmers: Hapmer generation if parental data is available
- Merqury: Completeness, consensus quality and phasing assessment
Synteny analysis
Annotation- GenomeTools gt gff3validator + FASTA/GFF correspondence: GFF3 validation
- GenomeTools gt stat: Annotation statistics
- GffRead, BUSCO: Gene-space completeness estimation in annotation proteins
- OrthoFinder: Phylogenetic orthology inference for comparative genomics
Refer to usage, parameters and output documents for details.
Note
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
Prepare an assemblysheet.csv file with following columns representing target assemblies and associated meta-data.
tag:A unique tag which represents the target assembly throughout the pipeline and in the final reportfasta:FASTA file
Now, you can run the pipeline using:
nextflow run plant-food-research-open/assemblyqc \
-revision <version> \
-profile <docker/singularity/.../institute> \
--input assemblysheet.csv \
--outdir <OUTDIR>Warning
Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters; see docs.
Download the pipeline to your /workspace/$USER folder. Change the parameters defined in the pfr/params.json file. Submit the pipeline to SLURM for execution.
sbatch ./pfr_assemblyqcplant-food-research-open/assemblyqc was originally written by Usman Rashid (@gallvp) and Ken Smith (@hzlnutspread).
Ross Crowhurst (@rosscrowhurst), Chen Wu (@christinawu2008) and Marcus Davy (@mdavy86) generously contributed their QC scripts.
Mahesh Binzer-Panchal (@mahesh-panchal) and Simon Pearce (@SPPearce) helped port the pipeline modules and sub-workflows to nf-core schema.
We thank the following people for their extensive assistance in the development of this pipeline:
The pipeline uses nf-core modules contributed by following authors:
If you would like to contribute to this pipeline, please see the contributing guidelines.
If you use plant-food-research-open/assemblyqc for your analysis, please cite it as:
AssemblyQC: A Nextflow pipeline for reproducible reporting of assembly quality.
Usman Rashid, Chen Wu, Jason Shiller, Ken Smith, Ross Crowhurst, Marcus Davy, Ting-Hsuan Chen, Ignacio Carvajal, Sarah Bailey, Susan Thomson & Cecilia H Deng.
Bioinformatics. 2024 July 30. doi: 10.1093/bioinformatics/btae477.
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
This pipeline uses code and infrastructure developed and maintained by the nf-core community, reused here under the MIT license.
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.