Identification and Discovery of Tumor Associated Microbes via RNAseq
Introduction
Pandora is a multi-step pipeline to find pathogen sequences in RNAseq data. It includes modules for host separation, assembly, blasting contigs, and orf discovery. As input, Pandora takes paired fastq files; as output, it produces a report.
Dependencies
The following programs must be in your PATH:
- python 2.7.x
- Samtools 1.4 (note: use older versions of samtools at your own risk - samtools is often not backwards compatible)
- STAR
- Bowtie2
- Trinity 2.1 (as of 2017, some newer versions of Trinity are plagued with bugs; note Trinity requires java and bowtie v1)
- BLAST 2.3.x
- featureCounts (Subread)
Pandora depends on the following Python modules:
- Biopython
- Pandas >= 0.20.1 (note: older versions of Pandas, such as 0.18.1, will throw an error)
- scipy
The exact list, with versions, is provided in the requirements.txt file.
And the best way to install these is the usual best practice of starting a virtualenv and running:
pip install -r requirements.txt
Workflow
To accomplish diverse tasks, Pandora has various subcommands (like, say, the program git).
The primary subcommand is scan, which is a pipeline comprised of the following steps:
- Subtraction of reads mapping to host genome
- De-Novo assembly of remaining reads
- BLAST of assembled contigs
- ORF search in contigs of unknown origin
- Filter and parse blast results into tidy human-readable report
Once you have multiple runs of scan, you can use the aggregate subcommand to make a report which collects statistics over your batch of runs. The aggregate subcommand also has multiple steps:
- Preprocess individual reports
- Generate aggregate report
Additional Files
Pandora requires various references and annotation files.
For scan step 1, please provide:
- a host genome indexed for STAR
- a host genome indexed for bowtie2
- (optional) a gtf describing the genes of the host
For scan step 3, please provide:
- the BLAST nucleotide collection nt database at ftp://ftp.ncbi.nlm.nih.gov/blast/db/
For scan step 4, you can optionally provide:
- the BLAST protein collection nr database at ftp://ftp.ncbi.nlm.nih.gov/blast/db/
For scan step 5, you can optionally provide:
- a text file of "blacklist" non-pathogen taxids for filtering. If you do not provide one, the script will use
resources/blacklist.txtby default. This list contains any taxid children of the nodes chordata (Taxonomy ID: 7711) or "other sequences" (Taxonomy ID: 28384) - the
names.dmpfile mapping taxID to names from ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/taxdump.tar.gz
Because there are a considerable number of files involved, you can specify their paths with a configuration file instead of command line flags.
See pandora.config.txt for example formatting.
Note that options specified as flags take precedence over options specified via the configuration file.
Scan: Usage Examples
pandora.py scan -id patient1 -r1 mate_1.fastq.gz -r2 mate_2.fastq.gz --gzip --refstar /path/ref/STAR --refbowtie /path/ref/bowtie/hg19 -db /path/ref/blastdb/nt --taxid2names /path/names.dmp
Here is an example command using a configuration file:
pandora.py scan -id patient1 -r1 mate_1.fastq.gz -r2 mate_2.fastq.gz --gzip --verbose -c pandora.config.txt
Keep intermediate files:
pandora.py scan -id patient1 -r1 mate_1.fastq.gz -r2 mate_2.fastq.gz --gzip --verbose -c pandora.config.txt --noclean
Run only steps 3 through 5:
pandora.py scan -id patient1 -r1 mate_1.fastq.gz -r2 mate_2.fastq.gz --gzip --verbose -c pandora.config.txt --steps 345
Example running pandora on AWS with Starcluster with an unmated read file:
mkdir -p logs; qsub -V -N pjob -e logs -o logs -S /usr/bin/python -cwd /opt/software/Pandora/pandora.py scan -id 1 --single --verbose --gzip -c /opt/software/Pandora/pandora.config.aws.txt -r1 single-end.fastq.gz --noclean --trinitycores 6 --trinitymem 30 --blast_threads 2
Note: the CUMC cluster and Starcluster on AWS behave differently.
You must use the --hpc flag to run on the CUMC hpc cluster.
Example:
pandora.py scan -id 1 --verbose --hpc --gzip -c pandora.config.hpc.txt -r1 mate_1.fastq.gz -r2 mate_2.fastq.gz
Scan: Output
Pandora produces three reports:
- report.contig.txt - a report keyed on contigs
- report.taxon.txt - a report keyed on taxids
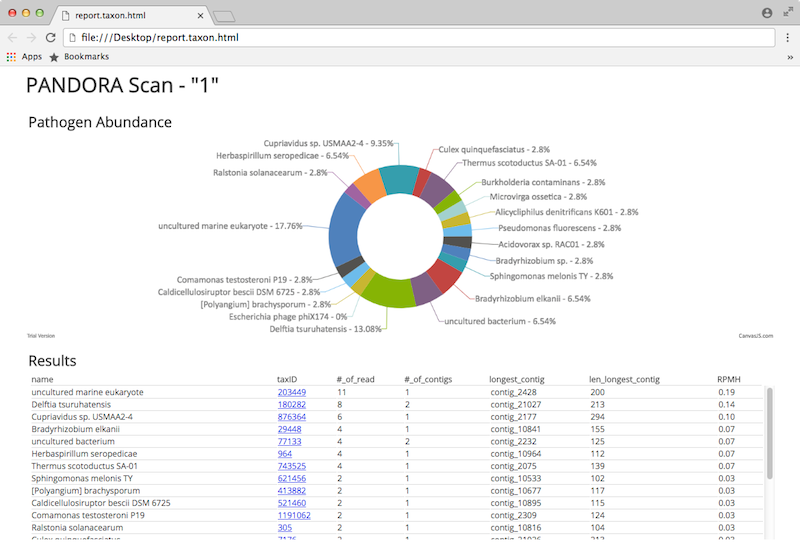
- report.taxon.html - a report for viewing in your browser
The later looks like this:
Docker
Because Pandora is a pipeline comprised of many programs, each with their own dependencies and input files, you may find it convenient to run Pandora via Docker.
One thing to be mindful of here is disk space and computing power. The mapping references, BLAST databases, etc. need approximately 100G of space. Additionally, some of the programs Pandora uses, such as Trinity, require significant computing power. We've tested input fastq files on the order of 100 megabytes zipped. We ran it on AWS with a t2.2xlarge instance (8 CPUs, 32G RAM) with 150G of disk space. Note we haven't gotten around to implementing parallelization in Docker, so it can be slow.
To use Docker, first copy the fordocker directory in this repository to some other location, where you intend to run it. For example:
cd Pandora
cp -R fordocker /home/ubuntu
The point here is to keep the code and output filepaths distinct. Next, enter your docker directory and make three new directories:
cd /home/ubuntu/fordocker
mkdir -p ref results data
Because Pandora requires some big files to run, they cannot be put into the Docker image without bloating it. For this reason, we use an alternate approach: downloading them locally into ref and mounting this folder as a Docker volume. Download the AWS CLI and you can grab the requisite files from The Rabadan Lab's S3 bucket:
cd ref
for i in GRCh37.75 pandora_resources taxdump nt; do
echo "***"${i};
aws s3 sync s3://ref-20170606/${i} ${i}/ ;
done
This takes care of the reference files. Next, rename your input files mate1.fq.gz and mate2.fq.gz (naturally, they should be zipped) and put them into data/. Docker will look for files with these names in this directory. (If you don't want to use this nomenclature, you can change the Dockerfile.)
Now it's time to build our docker image:
cd /home/ubuntu/fordocker
docker build -t pandoraslim .
Now we're ready to go. Run docker as:
docker run -v /home/ubuntu/fordocker/ref:/home/ref -v /home/ubuntu/fordocker/results:/home/results pandoraslim
The idea here is to pass our local ref and results directories to Docker. When Docker is finished running, the results will be in the results folder on our local computer (or instance). A note about the above command: the filepath /home/ubuntu/fordocker/ref may vary according to where you copy the fordocker directory; however, the /home/ref filepath is fixed.
If (for some reason) you want to run Docker interactively, you can do that as:
docker run -v /home/ubuntu/fordocker/ref:/home/ref -v /home/ubuntu/fordocker/results:/home/results -ti pandoraslim /bin/bash
Notes
Currently, Pandora makes use of the Oracle Grid Engine by default. The reason for this is that blast is computationally intensive, embarrassingly parallelizable, and lends itself very nicely to cluster computing. If you don't have access to a cluster, you can turn this off with the --noSGE flag (but blast will be slow).
Note that RNA-seq enriched for poly-A transcripts will miss prokaryotic pathogens.
Pipeline Status: Active Development