Associated repository for "Translation between Molecules and Natural Language" (EMNLP 2022).
Our paper has been accepted to EMNLP 2022. We will release a camera-ready version of the paper on arXiv soon. If you are using evaluation code, please see the README in ./evaluation
Table of Contents
- HuggingFace model checkpoints
- T5X-based model checkpoints
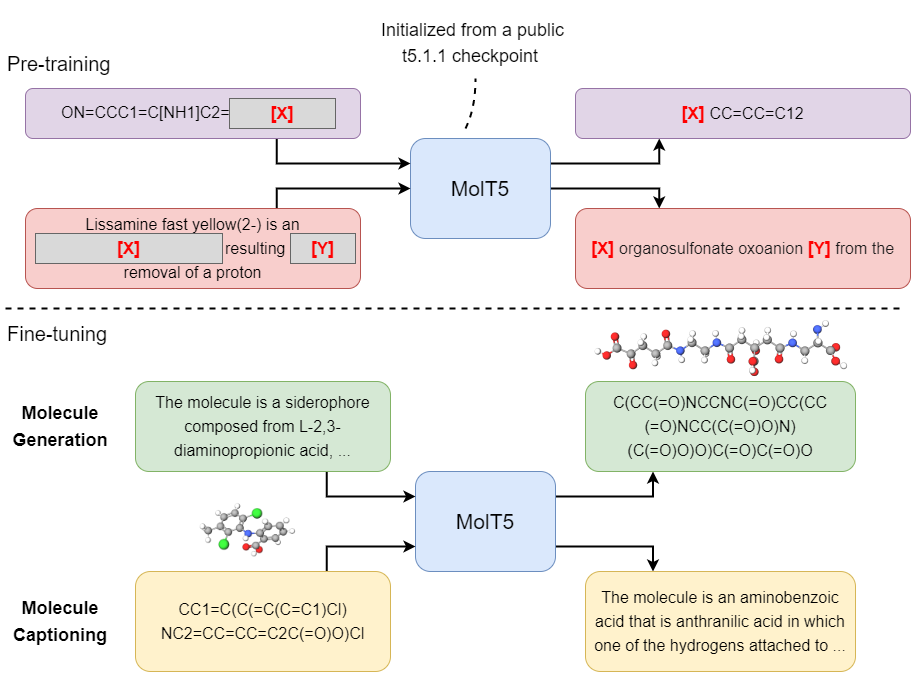
- Pretraining (MolT5-based models)
- Finetuning (MolT5-based models)
- Datasets
- Citation
All of our HuggingFace checkpoints are located here.
Pretrained MolT5-based checkpoints include:
- molt5-small (~77 million parameters)
- molt5-base (~250 million parameters)
- molt5-large (~800 million parameters)
You can also easily find our fine-tuned caption2smiles and smiles2caption models. For example, molt5-large-smiles2caption is a molt5-large model that has been further fine-tuned for the task of molecule captioning (i.e., smiles2caption).
Example usage for molecule captioning (i.e., smiles2caption):
from transformers import T5Tokenizer, T5ForConditionalGeneration
tokenizer = T5Tokenizer.from_pretrained("laituan245/molt5-large-smiles2caption", model_max_length=512)
model = T5ForConditionalGeneration.from_pretrained('laituan245/molt5-large-smiles2caption')
input_text = 'C1=CC2=C(C(=C1)[O-])NC(=CC2=O)C(=O)O'
input_ids = tokenizer(input_text, return_tensors="pt").input_ids
outputs = model.generate(input_ids, num_beams=5, max_length=512)
print(tokenizer.decode(outputs[0], skip_special_tokens=True))Example usage for molecule generation (i.e., caption2smiles):
from transformers import T5Tokenizer, T5ForConditionalGeneration
tokenizer = T5Tokenizer.from_pretrained("laituan245/molt5-large-caption2smiles", model_max_length=512)
model = T5ForConditionalGeneration.from_pretrained('laituan245/molt5-large-caption2smiles')
input_text = 'The molecule is a monomethoxybenzene that is 2-methoxyphenol substituted by a hydroxymethyl group at position 4. It has a role as a plant metabolite. It is a member of guaiacols and a member of benzyl alcohols.'
input_ids = tokenizer(input_text, return_tensors="pt").input_ids
outputs = model.generate(input_ids, num_beams=5, max_length=512)
print(tokenizer.decode(outputs[0], skip_special_tokens=True))We used the open-sourced t5x framework for pretraining MolT5-based models.
For pre-training MolT5-based models, please first go over this document. In our work, our pretraining task is a mixture of c4_v220_span_corruption and also our own task called zinc_span_corruption. The pretraining mixture is called zinc_and_c4_mix. The code snippet below illustrates how to define zinc_and_c4_mix (e.g., you can just add this code snippet to tasks.py). Our Gin config files for pretraining are located in configs/pretrain. Data files can be downloaded from here.
...
import tensorflow.compat.v2 as tf
...
seqio.TaskRegistry.add(
'zinc_span_corruption',
source=seqio.TFExampleDataSource(
split_to_filepattern={
'test': # Path to zinc_smiles_test.tfrecords,
'validation': # Path to zinc_smiles_val.tfrecords,
'train': # Path to zinc_smiles_train.tfrecords,
},
feature_description={
'text': tf.io.FixedLenFeature([], dtype=tf.string),
}),
preprocessors=[
functools.partial(
preprocessors.rekey, key_map={
'inputs': None,
'targets': 'text'
}),
seqio.preprocessors.tokenize,
preprocessors.span_corruption,
seqio.preprocessors.append_eos_after_trim,
],
output_features=DEFAULT_OUTPUT_FEATURES,
metric_fns=[])
seqio.MixtureRegistry.add('zinc_and_c4_mix', [('zinc_span_corruption', 1),
('c4_v220_span_corruption', 1)])
)We also used the t5x framework for finetuning MolT5-based models.
Please first go over this document.
Our Gin config files for finetuning are located in configs/finetune.
For each of the Gin file, you need to set the INITIAL_CHECKPOINT_PATH variables (please use one of the checkpoints mentioned in this section). Note that there are two new tasks, which are named caption2smiles and smiles2caption. The code snippet below illustrates how to define the tasks. Data files can be downloaded from here.
...
# Metrics
_TASK_EVAL_METRICS_FNS = [
metrics.bleu,
metrics.rouge,
metrics.sequence_accuracy
]
# Data Source
DATA_SOURCE = seqio.TFExampleDataSource(
split_to_filepattern={
'train': # Path to chebi_20_train.tfrecords,
'validation': # Path to chebi_20_dev.tfrecords,
'test': # Path to chebi_20_test.tfrecords
},
feature_description={
'caption': tf.io.FixedLenFeature([], dtype=tf.string),
'smiles': tf.io.FixedLenFeature([], dtype=tf.string),
'cid': tf.io.FixedLenFeature([], dtype=tf.string),
}
)
# Molecular Captioning (smiles2caption)
seqio.TaskRegistry.add(
'smiles2caption',
source=DATA_SOURCE,
preprocessors=[
functools.partial(
preprocessors.rekey,
key_map={
'inputs': 'smiles',
'targets': 'caption'
}),
seqio.preprocessors.tokenize,
seqio.preprocessors.append_eos_after_trim,
],
output_features=DEFAULT_OUTPUT_FEATURES,
metric_fns=_TASK_EVAL_METRICS_FNS,
)
# Molecular Captioning (caption2smiles)
seqio.TaskRegistry.add(
'caption2smiles',
source=DATA_SOURCE,
preprocessors=[
functools.partial(
preprocessors.rekey,
key_map={
'inputs': 'caption',
'targets': 'smiles'
}),
seqio.preprocessors.tokenize,
seqio.preprocessors.append_eos_after_trim,
],
output_features=DEFAULT_OUTPUT_FEATURES,
metric_fns=_TASK_EVAL_METRICS_FNS,
)If you found our work useful, please cite:
@article{edwards2022translation,
title={Translation between Molecules and Natural Language},
author={Edwards, Carl and Lai, Tuan and Ros, Kevin and Honke, Garrett and Ji, Heng},
journal={arXiv preprint arXiv:2204.11817},
year={2022}
}