This repository hosts Therapeutics Data Commons (TDC), an open, user-friendly and extensive machine learning dataset hub for therapeutics. So far, it includes more than 50+ datasets for 20+ tasks (ranging from target identification, virtual screening, QSAR to manufacturing, safety surveillance and etc) in many therapeutics development stages across small molecules and biologics.
- From Bench to Bedside: covers 50+ datasets for 20+ tasks in numerous therapeutics development stages across small molecules and biologics.
- User-friendly: 3 lines of codes to access any dataset and hassle-free installation.
- Ready-to-use: the dataset is processed into machine learning ready format.
- Data functions: TDC supports various useful functions such as data evaluators, realistic data split functions, data processing helpers, and molecule generation oracles!
- Benchmark: provides a benchmark for fair model comparison. A leaderboard will be released soon!
- Community-driven effort: TDC is a community-driven effort. Contact us if you want to contribute a new dataset or task!
To install TDC, simply open terminal and type:
pip install PyTDCThe core data loaders are designed to be lightweight, thus has minimum package dependency:
numpy
pandas
tqdm
scikit-learn
fuzzywuzzyFor other utilities requiring extra packages, TDC will print out the relevant installation instruction (e.g. for molecule generation oracles, TDC will print out RDKit installation instruction).
Note: TDC is in beta release. Please update your local copy regularly by
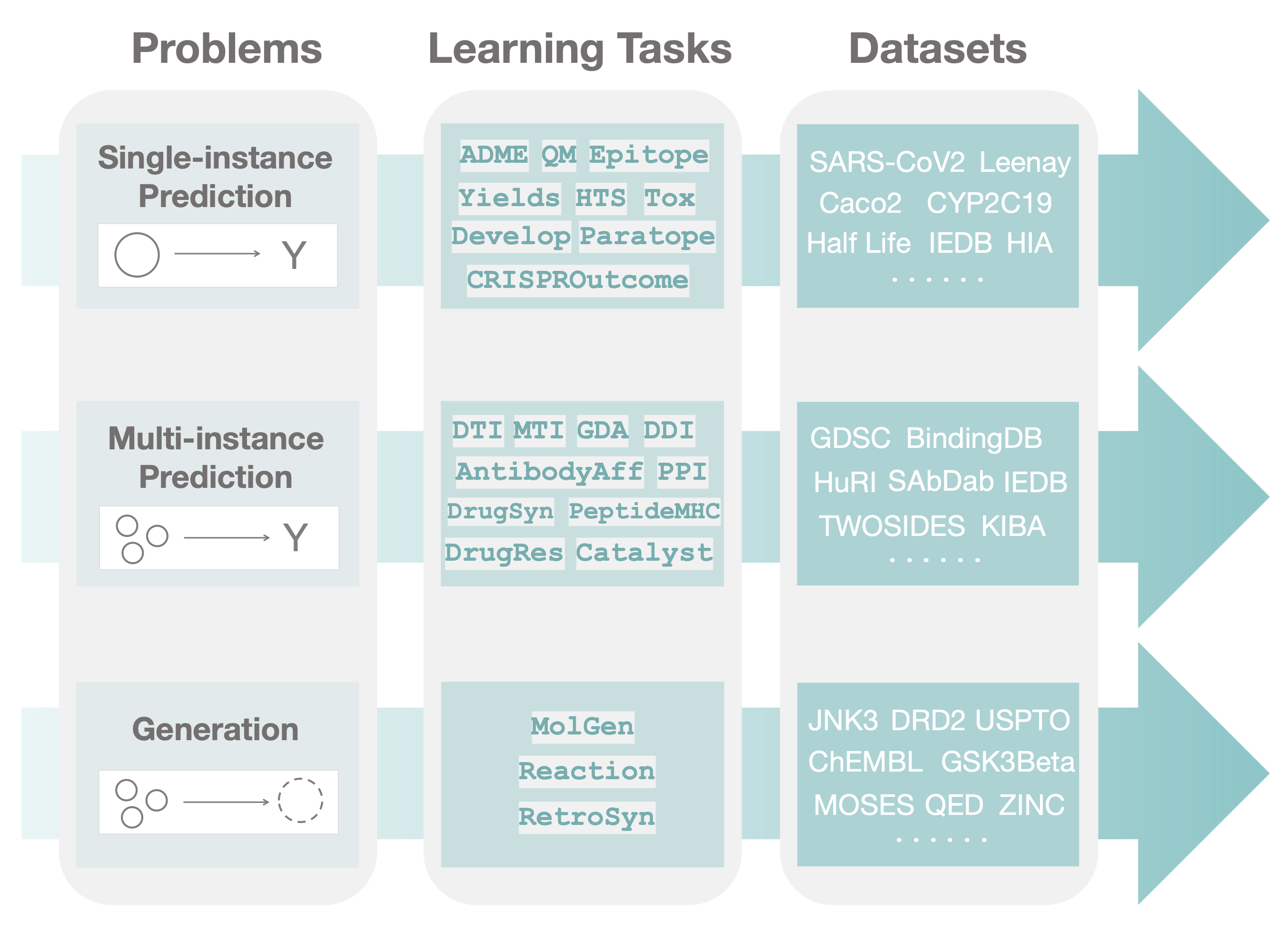
pip install PyTDC --upgradeTDC covers a wide range of therapeutics tasks with varying data structures. Thus, we organize it into three layers of hierarchies. First, we divide into three distinctive machine learning problems:
- Single-instance prediction
single_pred: Prediction of property given individual biomedical entity. - Multi-instance prediction
multi_pred: Prediction of property given multiple biomedical entities. - Generation
generation: Generation of new biomedical entity.
The second layer is task. Each therapeutic task falls into one of the machine learning problem. We create a data loader class for every task that inherits from the base problem data loader.
The last layer is dataset, where each task consists of many of them. As the data structure of most datasets in a task is the same, the dataset is used as a function input to the task data loader.
Supposed a dataset X is from therapeutic task Y with machine learning problem Z, then to obtain the data and splits, simply type:
from tdc.Z import Y
data = Y(name = 'X')
splits = data.split()For example, to obtain the HIA dataset from ADME therapeutic task in the single-instance prediction problem:
from tdc.single_pred import ADME
data = ADME(name = 'HIA_Hou')
# split into train/val/test using benchmark seed and split methods
split = data.get_split(method = 'scaffold', seed = 'benchmark')
# get the entire data in the various formats
data.get_data(format = 'df')Explore all therapeutic tasks and datasets in the website!
To retrieve the training/validation/test dataset split, you could simply type
data = X(name = Y)
data.get_split(seed = 'benchmark')
# {'train': df_train, 'val': df_val, ''test': df_test}You can specify the splitting method, random seed, and split fractions in the function by e.g. data.get_split(method = 'scaffold', seed = 1, frac = [0.7, 0.1, 0.2]). Check out the data split page on the website for details.
We provide various evaluation metrics for the tasks in TDC, which are described in model evaluation page on the website. For example, to use metric ROC-AUC, you could simply type
from tdc import Evaluator
evaluator = Evaluator(name = 'ROC-AUC')
score = evaluator(y_true, y_pred)We provide numerous data processing helper functions such as label transformation, data balancing, pair data to PyG/DGL graphs, negative sampling, database querying and so on. For individual function usage, please checkout the data processing page on the website.
For molecule generation tasks, we provide 10+ oracles for both goal-oriented and distribution learning. For detailed usage of each oracle, please checkout the oracle page on the website. For example, we want to retrieve the GSK3Beta oracle:
from tdc import Oracle
oracle = Oracle(name = 'GSK3B')
oracle(['CC(C)(C)....'
'C[C@@H]1....',
'CCNC(=O)....',
'C[C@@H]1....'])
# [0.03, 0.02, 0.0, 0.1]Note that the graph-to-graph paired molecule generation is provided as separate datasets.
If you found our work useful, please cite us:
@misc{tdc,
author={Huang, Kexin and Fu, Tianfan and Gao, Wenhao and Zhao, Yue and Zitnik, Marinka},
title={Therapeutics Data Commons: Machine Learning Datasets for Therapeutics},
howpublished={\url{http://tdc.hms.harvard.edu}},
month=nov,
year=2020
}
Paper is in progress and will come out soon.
We provide a series of tutorials for you to get started using TDC:
| Name | Description |
|---|---|
| 101 | Introduce TDC Data Loaders |
| 102 | Introduce TDC Data Functions |
| 103.1 | Walk through TDC Small Molecule Datasets |
| 103.2 | Walk through TDC Biologics Datasets |
| 104 | Generate 21 ADME ML Predictors with 15 Lines of Code |
| 105 | Molecule Generation Oracles |
We are actively working on the benchmark and leaderboard. We would release this feature in the next major release. In the meantime, if you have expertise or interest in helping build this feature, please send emails to us.
TDC is designed to be a community-driven effort. If you have new dataset or task or data function that wants to be included in TDC, please fill in this form!
Send emails to us or open an issue.
TDC is hosted in Harvard Dataverse. When dataverse is under maintenance, TDC will not able to retrieve datasets. Although rare, when it happens, please come back in couple of hours or check the status by visiting the dataverse website.
TDC is an open-source effort. Many datasets are aggregated from various public website sources. We use the Attribution-NonCommercial-ShareAlike 4.0 International license (open-source) to suffice many datasets requirement. If you feel there might be a potential infringement of the copyright, please let us know and we will address it ASAP.