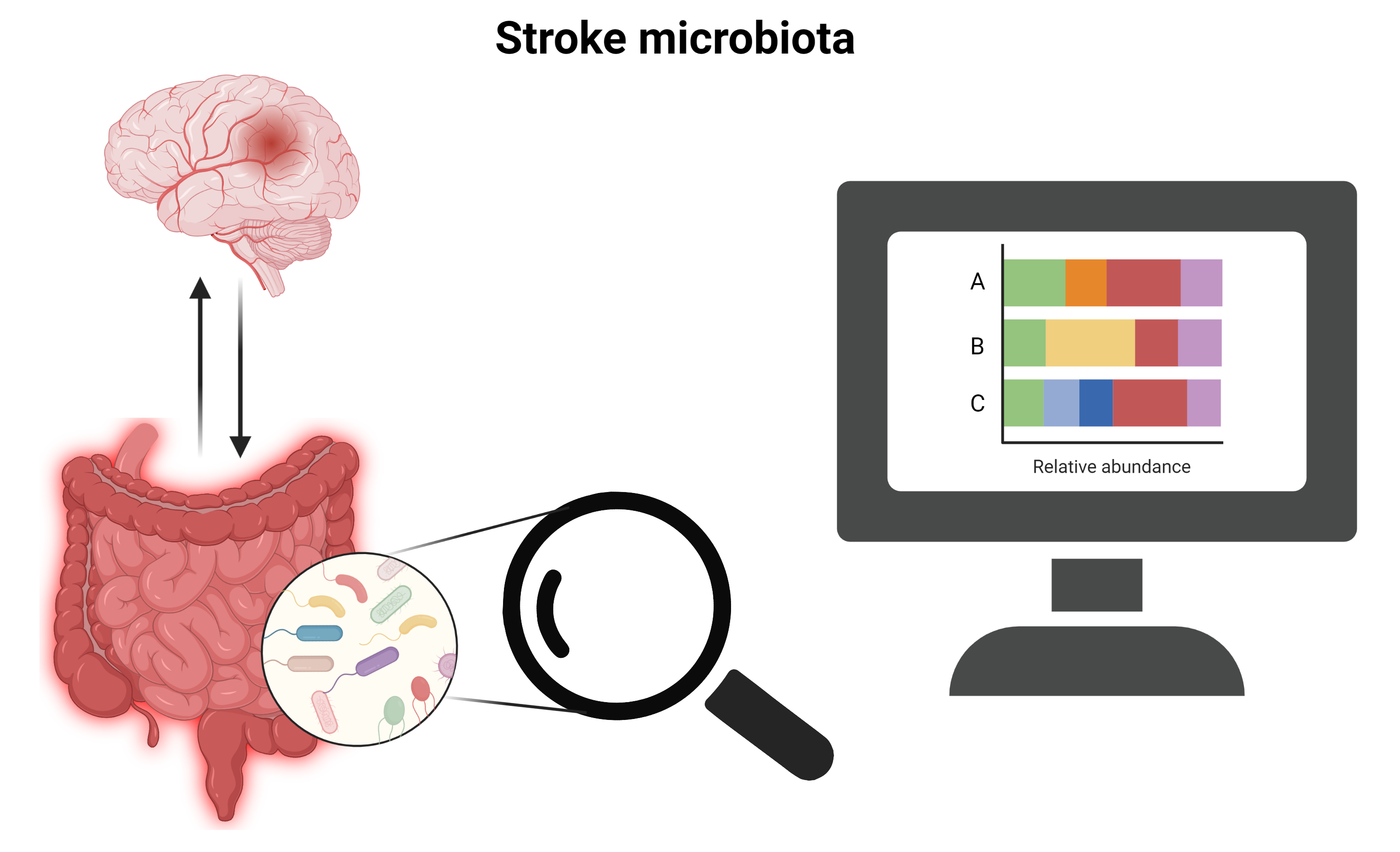
Data analysis part of the 2023 Neuroimmunolgical methods in stroke research at the ISD - Friday 17/02/23
Download and install the specific version of R for your OS here: (If using Windows 10/11, install Rtools as well)
Download and install Rstudio
Please check everything works before the course starts on Friday 17th.
Since some of you may not have any experience using R, we will very quickly go through some key concepts to help you understand better what we will doing in the analysis section later.
Download the course content and extract the folder.
Navigate to the course folder and open the "Neuroimmunolgical-methods-in-stroke-research-course-microbiota-analysis.Rmd" file in Rstudio.
Type and test these operations in the console in Rstudio
Addition
1 + 1
Subtraction
5 - 4
Multiplication
2 * 10
Division
9 / 3
Modulo
20 %/% 6
Square
4^2 or 4**2
Greater than
>
Less than
<
Equals
==
Does not equal
!=
There are 6 basic data types in R: logical, numeric, integer, character, raw and complex. Today (and in general) only the first four are important.
logical Can only have two values TRUE or FALSE (Shorthand:T,F).
numeric All numbers with or without decimal 100.34
integer Whole numbers 123L A number appended with L suffix denotes an integer in R
character Character or string values. Enclosed in either single 'microbe' or double "stroke" quotes.
Four most commonly used data structures
Vectors - ordered collection of data of a given length and all of the same type:
vector1 <- c(1,3,7,8,9)
vector2 <- c("A", "B", "C", "D")
print(vector2)Lists - ordered collection of data/objects of a given length. Can be composed of heterogenous types.
list1 <- list(vector1, vector2)
list2 <- list(1,2,3)
print(list1)Matrices - 2D data structure (rows x columns) of the same type
m <- matrix(c(1, 2, 3, 4, 5, 6, 7, 8, 9), nrow = 3, ncol = 3)
print(m)Dataframes - 2D heterogenous data structure (rows x columns)
df <- data.frame(bacteria = c("E. coli", "L. reuteri", "C. difficile"),
sample1 = c(10, 27, 61),
sample2 = c(9, 200, 43) )
print(df) Variables are assigned in R using the <- operator. A variable can be though of a a "box" you store a value in for later use.
They can be used to store a value which you want to be constant throughout your code e.g. a file name
x <- "Bacteria_data.txt" Or to save time by storing long strings/numbers
y <- 127575738398292929Objects such as vectors and dataframes can also be stored in variables as you saw above. df and vector2 are both variables.
As well as writing individual statements like we have done above we can also use logic to control the flow of our code, allowing us to repeat bits of code or only run if a given condition is met.
for loops allow us to repeat code a specified number of times e.g.
for (i in vector2){
print(i)
}This prints out each element in the vector2variable we defined earlier. Not particularly interesting though..
We will discover a more interesting use case later.
if/else statements allow us to control the flow of our code better:
x <- 100
y <- 10
if (x > y) {
print("x is higher")
} else if (x < y) {
print("y is higher")
}Change the values of x and y and see how the output changes.
Functions are used to abstract repetitive code in something which is reusable and modular, making code easier to read and debug.
Using the above if else example we could create a function called greater_than which tells us which value is highest in a more modular way
greater_than <- function(x, y) {
if (x > y) {
print("x is higher")
} else if (x < y) {
print("y is higher")
}
}greater_than(x=100, y=10)
greater_than(x=1, y=17)More interesting functions might perform a calculation for us and return the value
normalise <- function(x) {
x_norm <- t(100 * t(x) / colSums(x))
return(x_norm)
}Let's run this function on the matrix we created earlier to see what it does.
normalise(m)You can also use built-in functions or install packages to access functions written by others. It's usually the case that someone has already written a function for whatever you want to do. External packages can be downloaded using the install.packages function and loaded using the library function.
If you need help with a function you can also type ?functionname in the console e.g. ?log10 and the help for that function will show up, detailing what the function does, what inputs it expects and what value(s) it returns.
A few key concepts on loading and manipulating data.
Reading data
# install tidyverse
install.packages("tidyverse")
# load tidyverse
library(tidyverse)
# read in the palmer penguins csv file
penguins <- read_csv("penguins.csv")Manipulating data
Selecting columns
Base R
bill_length <- penguins$bill_length_mm
print(bill_length)
# alternative
bill_length <- penguins["bill_length_mm"]
print(bill_length)Tidyverse
# select certain columns
bill_length <- select(penguins, bill_length_mm)
print(bill_length)
# using pipe operator
bill_length <- penguins %>%
select(bill_length_mm)
print(bill_length)Filtering rows
Base R
gentoo <- penguins[penguins$species == "Gentoo", ]
print(gentoo)Tidyverse
gentoo <- filter(penguins, species == "Gentoo")
print(gentoo)
# with pipe
gentoo <- penguins %>%
filter(species == "Gentoo")
print(gentoo)Filtering and selecting in one with the pipe operator
gentoo_bill_length <- penguins %>%
filter(species == "Gentoo") %>%
select(bill_length_mm)
print(gentoo_bill_length)Now we understand a little bit of R we can move on to the actual analysis.
Here we will perform a very streamlined analysis of 16S rRNA sequencing data. This dataset comes from our lab and was published last year Sorbie et al, iScience 2022.
source("analysis_functions.R")Firstly, we will read the ASV table(s) and metadata into R, using phyloseq. We have included the data used in this publication to demonstrate our analysis pipeline.
ps <- import_as_pseq(asvtab = "data/ASV_seqtab_tax.tab",
mapping = "data/metadata.txt")Phyloseq is used as it provides a nice way of storing all the associated
data in one object or class. As you can see, the metadata and ASV
table are all combined here into ps. Importantly, each part can also
be accessed invidivually.
Here will transform the data using minimum sum normalisation and relative abundance for downstream analyses.
barplot(sort(sample_sums(ps)), horiz = TRUE, las = 2, xlab=NULL, main="Library sizes")
hist(sort(sample_sums(ps)), las = 2)Minimum sum normalisation works by dividing the sum in each sample by the minimum sum across all samples, thereby standardising each sample to a fixed sum.
Relative, transforms each sample into compositions, on a fixed scale of 0-100.
ps_norm <- transform(ps, transform = "mss")
ps_rel <- transform(ps, transform = "relative")We can see how this works by looking at the column sums:
colSums(otu_table(ps_rel))## RD118 RD122 RD124 RD126 RD127 RD128 RD130 RD131 RD132 RD162 RD165 RD166 RD167
## 100 100 100 100 100 100 100 100 100 100 100 100 100
## RD168 RD169 RD170 RD171 RD172 RD173 RD174
## 100 100 100 100 100 100 100
colSums(otu_table(ps_norm))## RD118 RD122 RD124 RD126 RD127 RD128 RD130 RD131 RD132 RD162 RD165
## 139635 139635 139635 139635 139635 139635 139635 139635 139635 139635 139635
## RD166 RD167 RD168 RD169 RD170 RD171 RD172 RD173 RD174
## 139635 139635 139635 139635 139635 139635 139635 139635 139635
Here we will calculate two different measures of alpha-diversity:
- Species richness, or the number of observed species
- Shannon effective diversity, measuring richness and evenness
The function calc_alpha wraps all of these calculations and only
requires the mss normalised phyloseq object as input, returning a
dataframe with a column for each above-mentioned dataframe.
alpha_div <- calc_alpha(ps_norm)Richness here is calculated as the the total number of observed species greater than 0.5 mss normalised abundance. This threshold is used to exclude noise (see Lagkouvardos et al 2017, PeerJ and Reitmeier et al 2021, ISME Communication for a more thorough explanation).
Shannon effective diversity is calculated as the exponent of the Shannon index:
This metric accounts for both the abundance and evenness of taxa.
Now we have calculated alpha diversity, we can merge this information with our metadata to generate a dataframe for plotting.
meta <-meta_to_df(ps_norm)
# merge alpha diversity df and metadata by rownames (0)
alpha_div_meta <- merge(alpha_div, meta, by=0)To plot the alpha diversity metrics we will use a boxplot with jittered
points layered on top. The function plot_boxplotwill do this for you,
we just need to set some parameters first.
Firstly, we will list the statistical comparisons we want to make, by
creating a list of vectors. In this case we only have two groups, stroke
and sham which can be written like: list(c("Stroke", "Sham")). If we
had an extra group, for example, “Control”, we would then have three
comparisons to define like so:
list(c("Control", "Sham"), c("Control", "Stroke"), c("Sham", "Stroke"))
Comparisons list
comps <- list(c("Stroke", "Sham"))We can also specify the colours we want to use in our plots here by creating a named vector of colours.
colours <- c("Sham" = "dodgerblue2", "Stroke" = "firebrick1")To generate the plot we need to provide the dataframe, the name of the
column containing the grouping variable (in this case simply “Group”),
the name of the column containing the values to plot (Richness here). To
colour by group we provide the column name of the grouping variable to
fill_var. We can then add the list of comparisons, some x and y-axis,
a title if desired and the plot colours.
In instances where the alphabetical order of your group levels does not
match the order you would like to plot them in, you can specify this
here with the group.orderparameter.
plot_boxplot(df = alpha_div_meta, variable_col = "Group", value_col = "Richness",
fill_var = "Group", comparisons_list = comps, xlab = "Group",
ylab = "Richness", p_title = "Richness Stroke vs Sham",
col_palette = colours, group.order = c("Sham", "Stroke")) ## [1] "Sham" "Stroke"
#### Shannon Effective
plot_boxplot(df = alpha_div_meta, variable_col = "Group", value_col = "Shannon.Effective",
fill_var = "Group", comparisons_list = comps, xlab = "Group",
ylab = "Shannon.Effective", p_title = "Shannon.Effective Stroke vs Sham",
col_palette = colours, group.order = c("Sham", "Stroke")) ## [1] "Sham" "Stroke"
Statistical significance is calculated internally in the plot_boxplot
function using unpaired wilcoxon, thus currently this function is only
suitable when groups are independent. Note that if no significance is
displayed on the plot, the the differences between groups were not
statistically significant.
Here we will calculate beta-diversity based on Generalized unifrac distance and plot an ordination of this using Non-metric multidimensional scaling.
The calc_betadiv function calculates a distance matrix, and an
ordination of that matrix, returning both as a list.
Various dissimilarity indices are available:
- Bray-Curtis - A count-based dissimilarity metric (beta-diversity), based on the fraction of overabundant counts.
- Unifrac - A phylogenetic beta-diversity metric measuring the fraction of unique branches in a phylogenetic tree.
- Weighted Unifrac - An extensions of unifrac taking abundance into account in addition
- Generalized Unifrac - A further extension of unifrac, placing less weight on highly-abundant lineages.
Similarly, there are also various ordination options:
- NMDS (default) - Non-Metric Multidimensional Scaling. An ordination method which attempts to represent the dissimilarity between samples, as closely as possible in a low-dimensional space.
- MDS/PCoA - Principal Coordinate analysis (also known as Metric Multidimensional Scaling). An ordination method which attempts to preserve distance between samples in a low dimensional Euclidean space.
betadiv <- calc_betadiv(ps_norm, dist = "bray", ord_method = "NMDS")## Run 0 stress 0.07932429
## Run 1 stress 0.07932448
## ... Procrustes: rmse 0.0005601283 max resid 0.001917895
## ... Similar to previous best
## Run 2 stress 0.08237485
## Run 3 stress 0.07932465
## ... Procrustes: rmse 0.0002580246 max resid 0.0008862056
## ... Similar to previous best
## Run 4 stress 0.08237496
## Run 5 stress 0.07957977
## ... Procrustes: rmse 0.01236273 max resid 0.04305789
## Run 6 stress 0.07932471
## ... Procrustes: rmse 0.0002874463 max resid 0.0009879163
## ... Similar to previous best
## Run 7 stress 0.07932445
## ... Procrustes: rmse 0.0001345411 max resid 0.0004614708
## ... Similar to previous best
## Run 8 stress 0.1209726
## Run 9 stress 0.08237489
## Run 10 stress 0.07932454
## ... Procrustes: rmse 0.0001872036 max resid 0.0006304013
## ... Similar to previous best
## Run 11 stress 0.08237504
## Run 12 stress 0.07932419
## ... New best solution
## ... Procrustes: rmse 0.000194693 max resid 0.0006719538
## ... Similar to previous best
## Run 13 stress 0.08237499
## Run 14 stress 0.0793245
## ... Procrustes: rmse 0.0003605691 max resid 0.001242011
## ... Similar to previous best
## Run 15 stress 0.08237502
## Run 16 stress 0.0823749
## Run 17 stress 0.08237499
## Run 18 stress 0.1210009
## Run 19 stress 0.3767472
## Run 20 stress 0.07932462
## ... Procrustes: rmse 0.0004240846 max resid 0.001456499
## ... Similar to previous best
## *** Solution reached
To plot beta diversity a convenience function plot-beta_div is
provided. We just need to provide the phyloseq object, the ordination
and distance matrix from the betadiv object above (This object is a list
so we can access elements with $ notation and their name), a grouping
variable and again the colours.
Since the ordination under the hood returns MDS as the column names, regardless of whether metric or non-metric dimensional scaling is used, we can adjust the axis labels by adding new x and y labels to our plot. All plotting functions return ggplot2 objects which can be added to and further customised as desired.
Within this function statistical testing of group separation is also carried out using the adonis function of vegan. This function performs a Permutational Multivariate Analysis of Variance or PERMANOVA test. The resulting R2 and p-value are added to the plot in the bottom left.
plot_beta_div(ps_norm, ordination = betadiv$Ordination,
dist_matrix = betadiv$Distance_Matrix, group_variable = "Group",
cols = colours)+
xlab("NMDS1") +
ylab("NMDS2")## Warning in plot_ordination(ps, ordination, color = group_variable): `Ordination
## species/OTU/taxa coordinate indices did not match `physeq` index names. Setting
## corresponding coordinates to NULL.
## Differential abundance
The final step of this pipeline is to calculate differentially abundant taxa between conditions.
This function performs the ancom-bc test, a compositionally aware differential abundance method and returns significant results. As input, only the phyloseq object and the column name of the grouping variable is required.
res_ancom <- ancom_da(ps_norm,formula="Group", group="Group")## Warning: The multi-group comparison will be deactivated as the group variable
## has < 3 categories
To visualise differentially abundant taxa, we provide a function which calculates fold change of significant taxa from above and plots diverging dot plot coloured by group, providing a clear figure showing which taxa change between conditions.
To this function, we need to provide the results of the ancom test
above, an ordered vector of the group levels e.g. c("Sham", "Stroke").
Additionally, we can provide the group colours to make interpretation
easier.
p <- plot_da(res_ancom, groups = c("Sham", "Stroke"), cols=colours)