An implementaton of probabilisitc principal components analysis which is a variant of vanilla PCA that can be used to
- compute factors where some of the data are missing
- interpolate data by using information from additional series
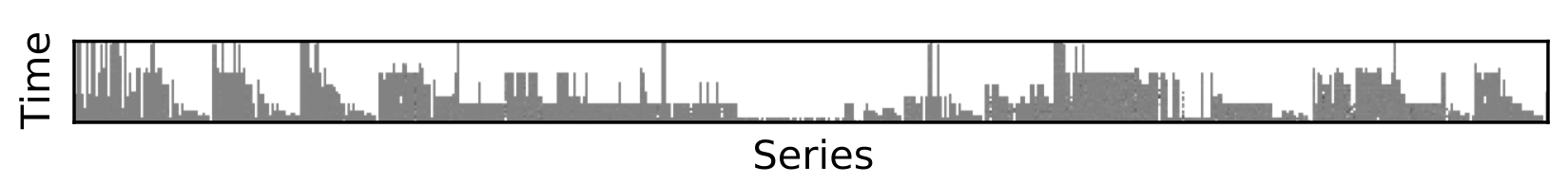
Often, you want to use PCA but your data is smattered with missing data. See below, where the white represents missing data in 14k+ time series in the Current Population Survey, a monthly survey of about 60k households conducted by the United States Census Bureau since 1940.
If enough of the data is not missing, you can fill in the missing data with sample means or some other interpolated value but if you have too much missing data, your rudimentary interpolation is going to overwhelm the signal in the data with noise. (Think about the limiting case with all but one missing data point).
A better way: suppose you had the latent factors representing the matrix. Construct a linear model for each series and then use the resulting model for interpolation. Intuitively, this will preserve the signal from the data as the interpolated values come from latent factors.
However, the problem is you never have these factors to begin with. The old chicken and egg problem. But no matter, fixed point algorithms via Probabilistic PCA to the rescue.
With this strategy, over 50 percent of the variance in those 14k+ time series in the CPS can be explained by just 12 factors.
Install via pip:
pip install ppca
Load in the data which should be arranged as n_samples by features. As usual, you should make sure your data is stationary (take first differences if possible) and standardized.
from ppca import PPCA
ppca = PPCA()
Fit the model with parameter d specifying the number of components and verbose printing convergence output if required.
ppca.fit(data=data, d=100, verbose=True)
The model parameters and components will be attached to the ppca object.
variance_explained = ppca.var_exp
components = ppca.data
model_params = ppca.C
If you want the principal components, call transform.
component_mat = ppca.transform()
Post fitting the model, save the model if you want.
ppca.save('mypcamodel')
Load a model, post instantiating a PPCA object. This will make fitting/transforming much faster.
ppca.load('mypcamodel.npy')