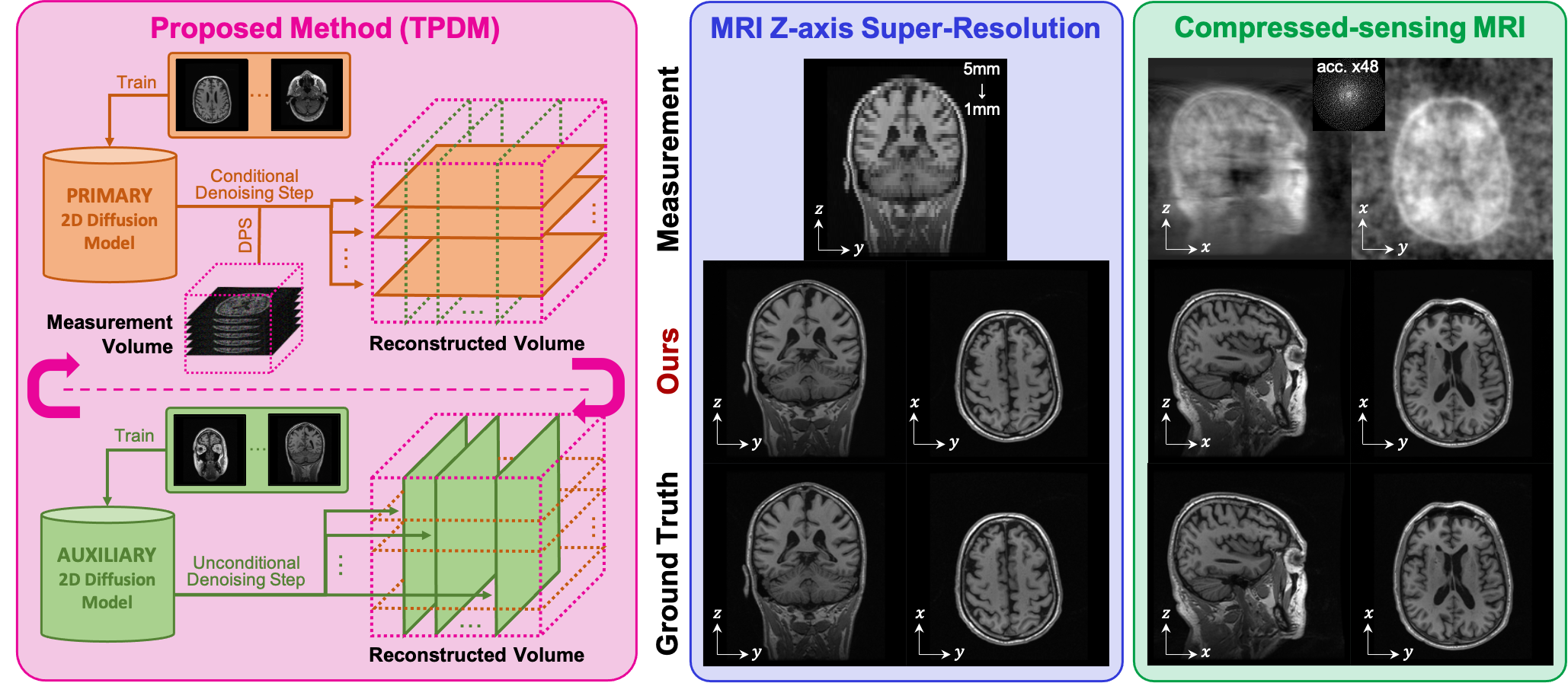
This repository TPDM is the official implementation of the paper Improving 3D Imaging with Pre-Trained Perpendicular 2D Diffusion Models (arxiv).
Proposed is a novel approach using two pre-trained 2D diffusion models perpendicular to each other to solve the 3D inverse problem effectively. Experimental results show its high effectiveness for 3D medical image reconstruction tasks, such as MRI Z-axis super-resolution, compressed sensing MRI, and sparse-view CT, generating high-quality voxel volumes for medical applications.
- 2024.04.21 We now additionally release checkpoints and sample volumes for the
LDCT-CUBEdataset. - 2024.04.21 The code now supports modifying the hyperparameter K introduced in the paper. Previously, it was fixed at K=2.
git clone https://github.com/hyn2028/tpdm.git
cd tpdmHere's a summary of the key dependencies.
- python 3.10
- pytorch 1.13.1
- CUDA 11.7
We highly recommend using conda to install all of the dependencies.
conda env create -f environment.yamlTo activate the environment, run:
conda activate tpdmDownload the pre-trained model for a 256x256x256 brain-MRI and abdominal-CT volume from the Google Drive link below. You can find detailed information about the dataset which we used in the paper.
| Dataset | Resolution | Model | Slice | Plane | Link |
|---|---|---|---|---|---|
| BMR-ZSR-1mm | 256 | primary model | coronal | YZ | link |
| auxiliary model | axial | XY | link | ||
| LDCT(AAPM)-CUBE | 256 | primary model | axial | XY | link |
| auxiliary model | coronal | YZ | link |
The
LDCT-CUBEcheckpoint originated from an experiment to observe the behavior of TPDM when the dataset was extremely small (10 volumes) to train a 3D model. Therefore, its PERFORMANCE may be INSUFFICIENT for use in abdominal CT-related tasks.
After downloading the checkpoints, place them in the ./checkpoints directory. The directory structure should look like this:
tpdm
├── checkpoints
│ ├── BMR_ZSR_256_XY
│ │ └── checkpoint.pth
│ ├── BMR_ZSR_256_YZ
│ │ └── checkpoint.pth
│ ├── AAPM_256_CUBE_SCLIP_XY
│ │ └── checkpoint.pth
│ └── AAPM_256_CUBE_SCLIP_YZ
│ └── checkpoint.pth
│ ...
Download sample brain-MRI and abdominal-CT volumes from the Google Drive link below.
| Dataset | Resolution | Slice | Plane | Link | Source |
|---|---|---|---|---|---|
| BMR-ZSR-1mm | 256 | coronal | YZ | link | synthetic |
| LDCT(AAPM)-CUBE | 256 | axial | XY | link | L067 volume from link |
NOTE: This sample brain-MRI volume is NOT A REAL MRI VOLUME. The pre-trained generative model generated it.
- Due to
BMR-ZSRdataset restrictions, we are unable to share the human subject test volumes we used in our experiments, so we provide a volume generated by unconditionalTPDMas a sample. - The sample volume of
LDCT-CUBEis the L067 volume processed from the AAPM 2016 Low Dose CT Grand Challenge dataset. The raw volume was converted to 256 cubes using the method mentioned in the paper. Afterwards, it was globally normalized such that the global signal (cube region only) ranged from -0.009425847 to 1.9688704.
After downloading sample volumes, place it in the ./dataset_sample directory. The directory structure should look like this:
tpdm
├── dataset_sample
│ ├── BMR_256_synthetic_0
│ │ ├── 000.npy
│ │ ├── 000.png
│ │ ...
│ │ ├── 255.npy
│ │ └── 255.png
│ └── AAPM_256_CUBE_SCLIP_XY_L067
│ ├── 000.npy
│ ├── 000.png
│ ...
│ ├── 255.npy
│ └── 255.png
│ ...
NOTE: The RESULT using the SYNTHETIC SAMPLE volume may be MUCH HIGHER than the reported .
- Note that the paper evaluated using real human subject MRI volumes, so reconstructions using the sample volumes provided may result in much higher results than those reported in the paper.
You can see the various arguments for all executables as shown below. There are many more arguments than the ones shown in the example. Additional arguments allow you to adjust the various hyperparameters such as the problem-related factor, DPS weight, and K.
python run_tpdm_uncond.py --help
python run_tpdm_mrzsr.py --help
python run_tpdm_csmri.py --help
python run_tpdm_svct.py --helpPlease don't rely on automatic DPS weights, as the optimal DPS weight can vary depending on the type of problem, parameters of the problem, measurements, and batch size. This is for reference only.
To generate a 256x256x256 brain-MRI volume with TPDM, run the following command:
python run_tpdm_uncond.py --batch-size <batch-size>To perform MR-ZSR on a sample volume with TPDM, run the following command:
python run_tpdm_mrzsr.py ./dataset_sample/BMR_256_synthetic_0 --batch-size <batch-size>To perform CS-MRI on a sample volume with TPDM, run the following command:
python run_tpdm_csmri.py ./dataset_sample/BMR_256_synthetic_0 --batch-size <batch-size>To perform SV-CT on a sample volume with TPDM, run the following command:
python run_tpdm_svct.py ./dataset_sample/AAPM_256_CUBE_SCLIP_XY_L067 --batch-size <batch-size>Our models were trained independently of the TPDM sampling procedure using score-SDE. Use the repository to train the two perpendicular diffusion models. Please refer to that repository for usage of that training code.
NOTE:
TPDM's sampling code is only implemented for variance-exploding SDE (VE-SDE).
See the ./configs/default_lsun_configs.py and ./configs/ve/BMR_ZSR_256.py for the configurations we used to train score-SDE model.
This code is based on score-SDE and its official implementation. We thank the authors for their work and for sharing the code.
If you find this repository useful in your research, please cite our paper:
@InProceedings{lee2023improving,
title={Improving 3D Imaging with Pre-Trained Perpendicular 2D Diffusion Models},
author={Lee, Suhyeon and Chung, Hyungjin and Park, Minyoung and Park, Jonghyuk and Ryu, Wi-Sun and Ye, Jong Chul}
booktitle={Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)},
month={October},
year={2023}
}