The current code was implemented using R v4.1.0, Bioconductor v3.13, Snakemake v5.5.0, and Python v3.6.8. All R dependencies (from GitHub, CRAN and Bioconductor) are listed under code/10-session_info.R and may be installed using the command contained therein.
config.yamlspecifies the R library and version to usecodecontains all R scripts used in the Snakemake workflowdatacontains raw, filtered and simulated scRNA-seq datasets,
as well as simulation parameter estimatesmetacontains two .json files that specify simulation method (methods.json) and reference subset (subsets.json) configurationsoutscontains all results from computations (typicallydata.frames) as .rds filesfigscontains all visual outputs as .pdf files, and correspondingggplotobjects as .rds files (for subsequent arrangement into 'super'-figures)
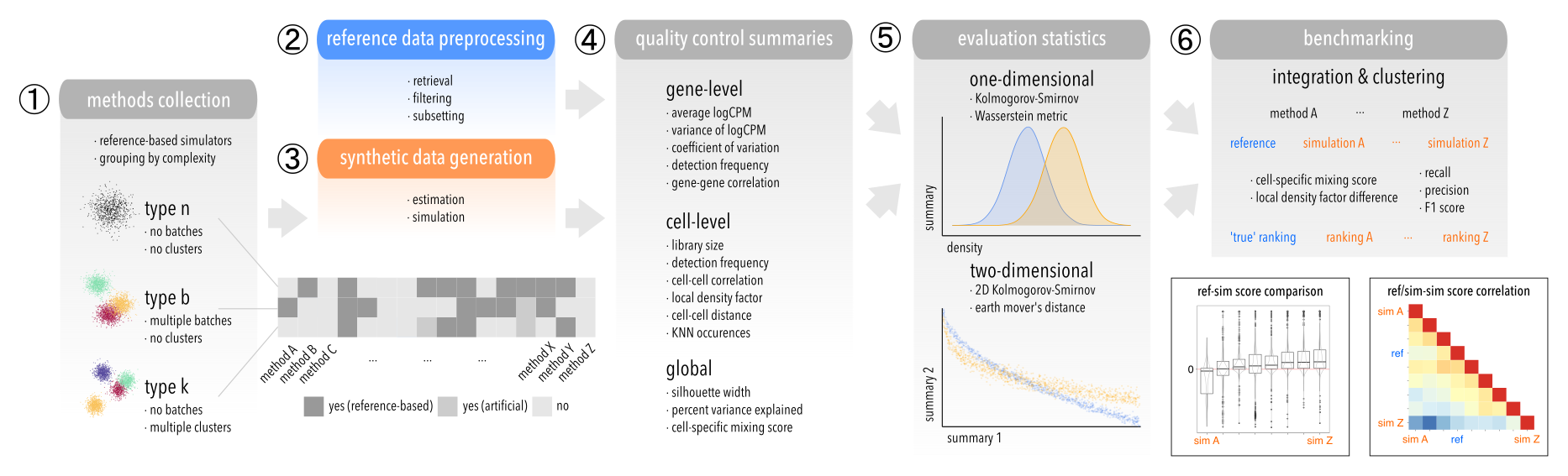
Simulation methods are tagged with one or many of the following labels, according to which scenario(s) they can accommodate:
nfor none: no clusters or batchesbfor batch: multiple batches, no clusterskfor cluster: multiple clusters, no batches
Similarly, we tag subsets (see below) with exactly one of these labels. This allows running each method on subsets they are capable of simulating.
1. Data retrieval
Each code/00-get_data-<datset_id>.R script retrieves a publicly available scRNA-seq dataset through from which a SingleCellExperiment is constructed and written to data/00-raw/<datset_id>.rds
2. Filtering
code/01-fil_data.R is applied to each raw dataset as to
- remove batches, cluster, or batch-cluster instances with fewer than 50 cells (depending on the dataset's complexity)
- keep genes with a count of at least 1 in at least 10 cells, and remove cells with fewer than 100 detected genes
Filtered data are written to data/01-fil/<datset_id>.rds.
3. Subsetting
Because different methods can accommodate only some features (e.g. multiple batches or clusters, both or neither), code/02-sub_data.R creates specific subsets in data/02-sub/<datset_id>.<subset_id>,rds. We term these ref(erence)sets (i.e. <datset_id>.<subset_id> = <refset_id>), as they serve as the input reference data for simulation.
1. Parameter estimation
Simulation parameters are estimated with code/03-est_pars.R, which in term sources a code/03-est_pars-<method_id>.R script that executes a method's parameter estimation function(s). In cases where no separate estimation takes place, this returns NULL. Parameter estimates for each combination of <refset_id.<method_id> = <simset_id> are written to data/04-est/<simset_id>.rds.
2. Data simulation
Data is simulated with code/04-sim_data.R, which in term sources a code/04-sim_data-<method_id>.R script that executes a method's simulation function. Simulations for each combination of <refset_id> and method_id are written to data/05-sim/<refset_id>,<method_id>.rds.
Various quality control (QC) summaries are computed with code/05-calc_qc.R, which in term sources a set of code/05-calc_qc-<metric_id>.R scripts. QC results for reference and simulated data are written to outs/qc_ref-<refset_id>,<metric_id>.rds and outs/qc_sim-<simset_id>,<metric_id>.rds, respectively. At current, we consider:
1. Gene-level
frq: detection frequency (i.e., fraction of cells with non-zero counts)avg/var: average/variance of logCPMcv: coefficient of variationcor: gene-to-gene correlation
2. Cell-level
frq: detection frequency (i.e., fraction of genes with non-zero counts)lls: log-transformed library size (total counts)cor: cell-to-cell correlationpcd: cell-to-cell distance (in PCA space)knn: number of KNN occurrencesldf: local density factor
3. Global
sw: Silhouette width (using batch/cluster labels as classes)cms: cell-specific mixing score (using batch/cluster labels as batches)pve: percent variance explained (of gene expression = logCPM, by batch/cluster)
Noteworthily, we compute each summary for different groupings of cells (depending on the dataset's complexity):
- globally, i.e. across all cells
- at the batch-level, i.e. for each batch
- at the cluster-level, i.e. for each cluster
Global summaries are computed at the batch-/cluster-level only, as they require a grouping variable.
We compare summaries between reference and simulated data in both one- (code/06-stat_1d.R) and two-dimensional settings (code/06-statl_2d.R). For the latter, every combination of gene- and cell-level metrics is considered, excluding correlations and global summaries. Furthermore, metrics are evaluated for each cell grouping, i.e. we perform a test globally, for each batch and cluster (again, depending on the dataset's complexity). Test results are written to outs/stat_1d,<refset_id>,<metric_id>,<stat1d_id>.rds for 1D, and outs/stat_2d,<refset_id>,<metric1_id>,<metric2_id>,<stat2d_id>.rds for 2D tests.
1. One-dimensional
- Kolmogorov-Smirnov (KS) test
- Wasserstein metric
2. Two-dimensional
- two-dimensional KS test
- Earth Mover's Distance (EMD)
Each 05-calc_batch-x.R script wraps around an integration method that is applied in 05-calc_batch.R to the set of type b subsets. The output corrected assay data or integrated cell embeddings (depending on the method) are written to outs/batch_ref/sim-<ref/simset_id>,<batch_method>.rds for every reference and simulation, respectively. Results are evaluated by 06-eval_batch.R, which computes the following set of metrics:
- cell-specific mixing score (CMS)
- difference in local density factor ($\Delta$LDF)
- batch correction score (BCS)
Each 05-calc_clust-x.R script wraps around an integration method that is applied in 05-calc_clust.R to the set of type b subsets. The output cluster assignments are written to outs/clust_ref/sim-<ref/simset_id>,<clust_method>.rds for every reference and simulation, respectively. Results are evaluated by 06-eval_clust.R, which computes the following set of metrics:
- precision (P) and recall (R)
- F1 score (harmonic mean of P and R)
Finally, results are collected across refset_ids and method_ids (jointly or separated by type), and visualized in various ways using as set of 07-plot_x.R scripts. Output figures are written to plts as .pdf files, along with the corresponding ggplot objects as .rds files. Lastly, 08-fig_x.R scripts are used to combined various ggplots into figures that are saved to figs as .pdf files.
In principle, any dataset for which a code/00-get_data-<dataset_id>.R script exists will be accessible to the workflow. However, data will only be retrieved if the dataset appears in meta/subsets.json. Hence,
To exclude a dataset from the workflow, i) (re)move the corresponding code/00-get_data-<dataset_id>.R script; or, ii) remove or comment out any associated meta/subsets.json entries.
Similarly, a new dataset can be added by supplying an adequate code/00-get_data-<dataset_id>.R script, and adding an entry to the meta/subsets.json configuration that specifies the subset ID, the number of genes/cells to sample (NULL for all), which batch(es)/cluster(s) to retain, as well as the resulting subset's type (one of n,b,k,g).
The Snakemake will automatically include any simulation method for which a code/03-est_pars-<method_id>.R and code/04-sim_data-<method_id>.R script exists. Secondly, meta/methods.json will determine on which type(s) of dataset(s) each method should be run. Thus,
To exclude a method from the workflow, either i) set "<method_id>": "x" in the meta/methods.json file (or anything other than n,b,k,g); or, ii) (re)move the parameter estimation and/or simulation script from the code directory.
Analogous to the above, adding a method to the benchmark requires i) adding a code/03-est_pars-<method_id>.R and code/04-sim-data-<method_id>.R script; and, ii) adding an entry for the method_id to the meta/methods.json file. Importantly, the R script for parameter estimation should handle batches (colData column batch), clusters (colData column cluster), both or neither. And the method's type(s) should be specified accordingly (n for neither, b/k for batches/clusters, g for groups), e.g. "<method_id>": ["n", "k"] for a method that supports 'singular' datasets, as well as ones with multiple clusters.