We can use the function plt_summary() in
rcode/00_pkg_functions.R to plot the
records, overlay with grid cells that we can specify resolution, and
count the number of cells with enough data.
d = readr::read_csv("data/Claytonia virginica_inat.csv") %>%
dplyr::select(longitude, latitude, everything()) %>%
filter(flowers == 1) %>%
drop_na(longitude, latitude) %>%
rename(id_iNat = id)
#> Parsed with column specification:
#> cols(
#> id = col_double(),
#> flowers = col_double(),
#> scientific_name = col_character(),
#> observed_on = col_date(format = ""),
#> latitude = col_double(),
#> longitude = col_double()
#> )
#> Warning: 15 parsing failures.
#> row col expected actual file
#> 2118 flowers a double U 'data/Claytonia virginica_inat.csv'
#> 2308 flowers a double U 'data/Claytonia virginica_inat.csv'
#> 2477 flowers a double U 'data/Claytonia virginica_inat.csv'
#> 2524 flowers a double U 'data/Claytonia virginica_inat.csv'
#> 2600 flowers a double U 'data/Claytonia virginica_inat.csv'
#> .... ....... ........ ...... ...................................
#> See problems(...) for more details.
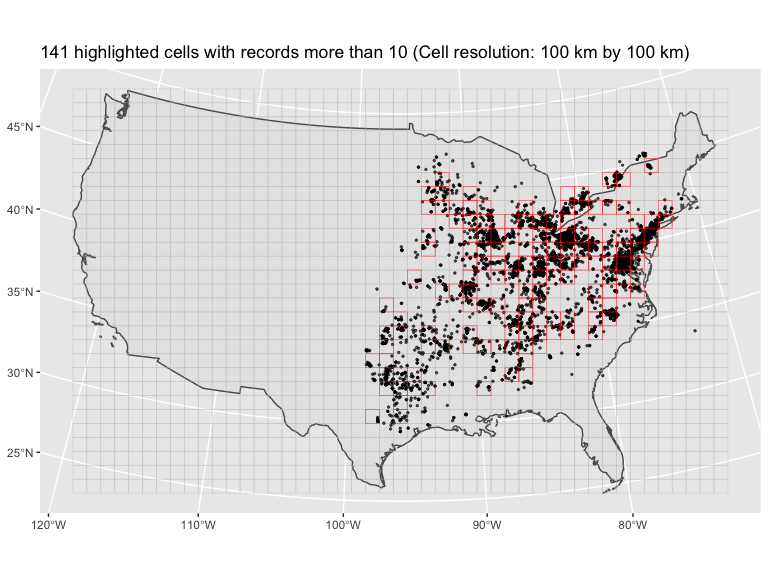
cell_100k = plt_summary(cell_size = 100000, dat = d, n_per_cell = 10)
#> 141 cells with records more than 10To get the grid cells with enough records specified by n_per_cell
argument:
filter(cell_100k$cells_with_data, enough_data)
#> Simple feature collection with 141 features and 5 fields
#> geometry type: POLYGON
#> dimension: XY
#> bbox: xmin: -253235.9 ymin: -830092.5 xmax: 1946764 ymax: 1069908
#> epsg (SRID): NA
#> proj4string: +proj=aea +lat_1=29.5 +lat_2=45.5 +lat_0=37.5 +lon_0=-96 +x_0=0 +y_0=0 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs
#> First 10 features:
#> id_cells n enough_data geometry long_cell lat_cell
#> 1 257 11 TRUE POLYGON ((-253235.9 -830092... -98.12156 30.49581
#> 2 259 19 TRUE POLYGON ((-53235.92 -830092... -96.03379 30.51622
#> 3 306 12 TRUE POLYGON ((-53235.92 -730092... -96.03416 31.41551
#> 4 352 59 TRUE POLYGON ((-153235.9 -630092... -97.10201 32.30789
#> 5 353 25 TRUE POLYGON ((-53235.92 -630092... -96.03454 32.31325
#> 6 354 22 TRUE POLYGON ((46764.08 -630092.... -94.96707 32.30854
#> 7 355 13 TRUE POLYGON ((146764.1 -630092.... -93.89985 32.29375
#> 8 359 16 TRUE POLYGON ((546764.1 -630092.... -89.63894 32.13401
#> 9 399 54 TRUE POLYGON ((-153235.9 -530092... -97.11453 33.20422
#> 10 400 20 TRUE POLYGON ((-53235.92 -530092... -96.03494 33.20963To get the raw records that to be used (i.e. fall within cells with enough data):
cell_100k$dat_to_use
#> Simple feature collection with 8173 features and 5 fields
#> geometry type: POINT
#> dimension: XY
#> bbox: xmin: -246930.5 ymin: -827406.3 xmax: 1944863 ymax: 1025360
#> epsg (SRID): NA
#> proj4string: +proj=aea +lat_1=29.5 +lat_2=45.5 +lat_0=37.5 +lon_0=-96 +x_0=0 +y_0=0 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs
#> # A tibble: 8,173 x 6
#> id_iNat flowers scientific_name observed_on geometry id_cells
#> * <dbl> <dbl> <chr> <date> <POINT [m]> <int>
#> 1 58129 1 Claytonia virginica 2012-03-14 (1127061 268941.6) 740
#> 2 60878 1 Claytonia virginica 2012-03-18 (1607161 159337.7) 698
#> 3 62915 1 Claytonia virginica 2012-04-06 (1199452 498004.4) 882
#> 4 71851 1 Claytonia virginica 2012-04-29 (1139628 458921) 834
#> 5 229730 1 Claytonia virginica 2013-04-05 (1514004 -38586.48) 603
#> 6 236518 1 Claytonia virginica 2013-04-14 (1179408 485569.1) 882
#> 7 240011 1 Claytonia virginica 2013-04-18 (1201700 497737.7) 882
#> 8 247008 1 Claytonia virginica 2013-04-27 (1309953 383145.3) 836
#> 9 249102 1 Claytonia virginica 2013-04-29 (1169172 491358.7) 882
#> 10 465413 1 Claytonia virginica 2010-04-10 (676948.7 532757.4) 877
#> # … with 8,163 more rowsTo see the plot:
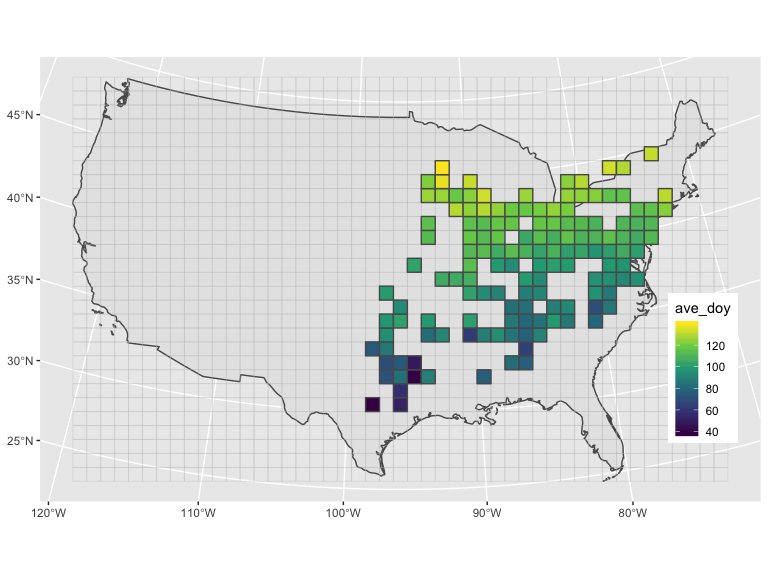
cell_100k$figTo plot the summarized data of 2019:
cell_100k$fig_base +
geom_sf(data = mutate(cell_100k$dat_to_use, observed_yr = lubridate::year(observed_on),
observed_doy = lubridate::yday(observed_on)) %>%
st_drop_geometry() %>%
filter(observed_yr %in% c(2019)) %>%
group_by(observed_yr, id_cells) %>%
summarise(ave_doy = mean(observed_doy, na.rm = T)) %>%
left_join(cell_100k$grids, by = "id_cells") %>%
st_sf(),
aes(fill = ave_doy)) +
scale_fill_viridis_c() +
theme(legend.position = c(0.92, 0.3))