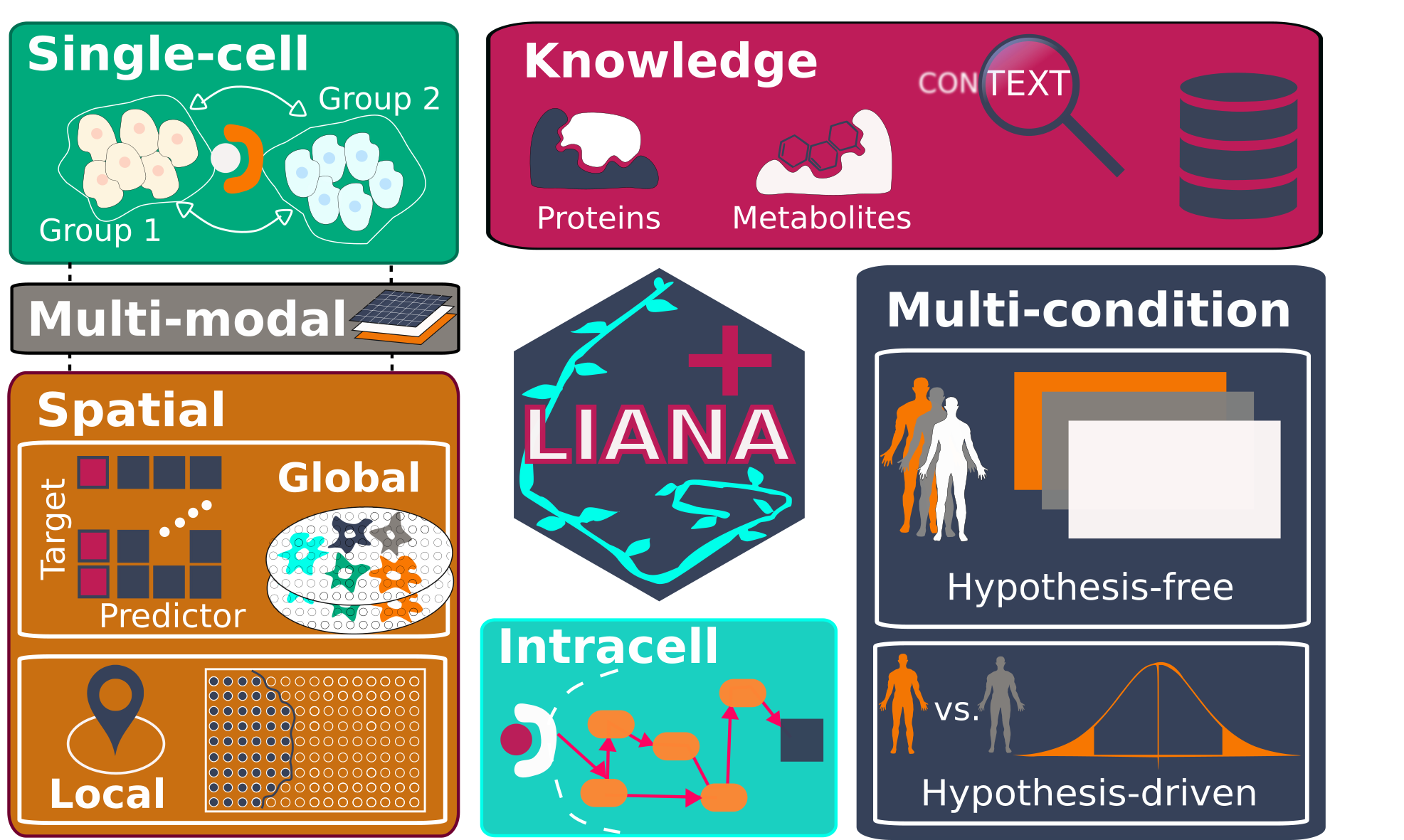
LIANA+ is an efficient framework that integrates and extends existing methods and knowledge to study cell-cell communication in single-cell, spatially-resolved, and multi-modal omics data.
LIANA+ is currently at it's alpha stage - i.e. we are actively working on improving it.
The API visible to the user should be stable, specifically liana.method.sc, but some differences in terms of secondary parameters, or default settings are possible.
More importantly, we welcome suggestions, ideas, and contributions! Please use do not hesitate to contact us, or use the issues or the LIANA+ Development project to make suggestions.
- LIANA's basic tutorial in dissociated single-cell data
-
Learn spatially-informed relationships with MISTy across (multi-)views
-
Estimate local spatially-informed bivariate metrics with LIANA's bivariate scores
-
Hypothesis-testing for CCC with PyDeSeq2 that also shows the inference of causal intracellular signalling networks
-
Multicellular programmes with MOFA. Using MOFA to obtain coordinates gene expression programmes across samples and conditions, as done in Ramirez et al., 2023
-
LIANA with MOFA. Using MOFA to infer intercellular communication programmes across samples and conditions, as initially proposed by cell2cell-Tensor
-
LIANA with cell2cell-Tensor to extract intercellular communication programmes across samples and conditions. Extensive tutorials combining LIANA & cell2cell-Tensor are available here.
- We also refer users to the Cell-cell communication chapter in the best-practices guide from Theis lab. There we provide an overview of the common limitations and assumptions in CCC inference from (dissociated single-cell) transcriptomics data.
For further information please check LIANA's API documentation.
Dimitrov, D. ... Saez-Rodriguez, J.
Dimitrov, D., Türei, D., Garrido-Rodriguez M., Burmedi P.L., Nagai, J.S., Boys, C., Flores, R.O.R., Kim, H., Szalai, B., Costa, I.G., Valdeolivas, A., Dugourd, A. and Saez-Rodriguez, J. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat Commun 13, 3224 (2022). https://doi.org/10.1038/s41467-022-30755-0
Similarly, please consider citing any of the methods and/or resources implemented in liana, that were particularly relevant for your research!