GPL is an unsupervised domain adaptation method for training dense retrievers. It is based on query generation and pseudo labeling with powerful cross-encoders. To train a domain-adapted model, it needs only the unlabeled target corpus and can achieve significant improvement over zero-shot models.
For more information, checkout our publication:
For reproduction, please refer to this snapshot branch.
One can either install GPL via pip
pip install gplor via git clone
git clone https://github.com/UKPLab/gpl.git && cd gpl
pip install -e .Meanwhile, please make sure the correct version of PyTorch has been installed according to your CUDA version.
GPL accepts data in the BeIR-format. For example, we can download the FiQA dataset hosted by BeIR:
wget https://public.ukp.informatik.tu-darmstadt.de/thakur/BEIR/datasets/fiqa.zip
unzip fiqa.zip
head -n 2 fiqa/corpus.jsonl # One can check this data format. Actually GPL only need this `corpus.jsonl` as data input for training.Then we can either use the python -m function to run GPL training directly:
export dataset="fiqa"
python -m gpl.train \
--path_to_generated_data "generated/$dataset" \
--base_ckpt "distilbert-base-uncased" \
--gpl_score_function "dot" \
--batch_size_gpl 32 \
--gpl_steps 140000 \
--new_size -1 \
--queries_per_passage -1 \
--output_dir "output/$dataset" \
--evaluation_data "./$dataset" \
--evaluation_output "evaluation/$dataset" \
--generator "BeIR/query-gen-msmarco-t5-base-v1" \
--retrievers "msmarco-distilbert-base-v3" "msmarco-MiniLM-L-6-v3" \
--retriever_score_functions "cos_sim" "cos_sim" \
--cross_encoder "cross-encoder/ms-marco-MiniLM-L-6-v2" \
--qgen_prefix "qgen" \
--do_evaluation \
# --use_amp # Use this for efficient training if the machine supports AMP
# One can run `python -m gpl.train --help` for the information of all the arguments
# To reproduce the experiments in the paper, set `base_ckpt` to "GPL/msmarco-distilbert-margin-mse" (https://huggingface.co/GPL/msmarco-distilbert-margin-mse)or import GPL's trainining method in a python script:
import gpl
dataset = 'fiqa'
gpl.train(
path_to_generated_data=f"generated/{dataset}",
base_ckpt="distilbert-base-uncased",
# base_ckpt='GPL/msmarco-distilbert-margin-mse',
# The starting checkpoint of the experiments in the paper
gpl_score_function="dot",
# Note that GPL uses MarginMSE loss, which works with dot-product
batch_size_gpl=32,
gpl_steps=140000,
new_size=-1,
# Resize the corpus to `new_size` (|corpus|) if needed. When set to None (by default), the |corpus| will be the full size. When set to -1, the |corpus| will be set automatically: If QPP * |corpus| <= 250K, |corpus| will be the full size; else QPP will be set 3 and |corpus| will be set to 250K / 3
queries_per_passage=-1,
# Number of Queries Per Passage (QPP) in the query generation step. When set to -1 (by default), the QPP will be chosen automatically: If QPP * |corpus| <= 250K, then QPP will be set to 250K / |corpus|; else QPP will be set 3 and |corpus| will be set to 250K / 3
output_dir=f"output/{dataset}",
evaluation_data=f"./{dataset}",
evaluation_output=f"evaluation/{dataset}",
generator="BeIR/query-gen-msmarco-t5-base-v1",
retrievers=["msmarco-distilbert-base-v3", "msmarco-MiniLM-L-6-v3"],
retriever_score_functions=["cos_sim", "cos_sim"],
# Note that these two retriever model work with cosine-similarity
cross_encoder="cross-encoder/ms-marco-MiniLM-L-6-v2",
qgen_prefix="qgen",
# This prefix will appear as part of the (folder/file) names for query-generation results: For example, we will have "qgen-qrels/" and "qgen-queries.jsonl" by default.
do_evaluation=True,
# use_amp=True # One can use this flag for enabling the efficient float16 precision
)One can also refer to this toy example on Google Colab for better understanding how the code works.
The workflow of GPL is shown as follows:
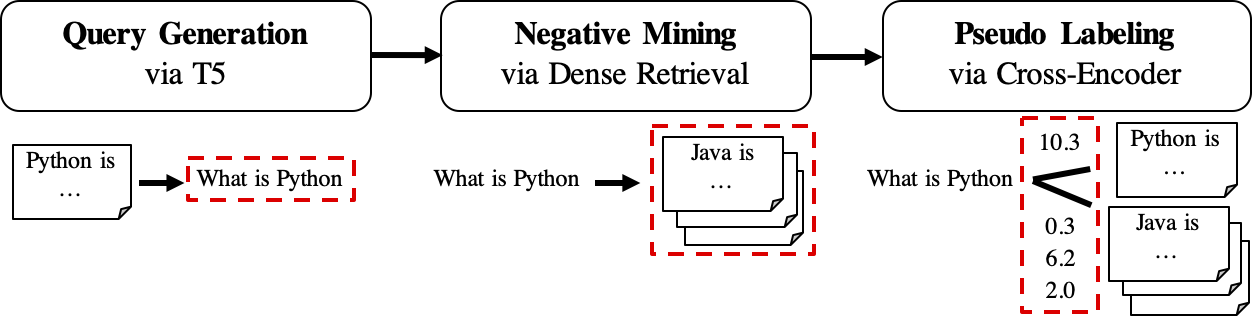
- GPL first use a seq2seq (we use BeIR/query-gen-msmarco-t5-base-v1 by default) model to generate
queries_per_passagequeries for each passage in the unlabeled corpus. The query-passage pairs are viewed as positive examples for training.Result files (under path
$path_to_generated_data): (1)${qgen}-qrels/train.tsv, (2)${qgen}-queries.jsonland also (3)corpus.jsonl(copied from$evaluation_data/); - Then, it runs negative mining with the generated queries as input on the target corpus. The mined passages will be viewed as negative examples for training. One can specify any dense retrievers (SBERT or Huggingface/transformers checkpoints, we use msmarco-distilbert-base-v3 + msmarco-MiniLM-L-6-v3 by default) or BM25 to the argument
retrieversas the negative miner.Result file (under path
$path_to_generated_data): hard-negatives.jsonl; - Finally, it does pseudo labeling with the powerful cross-encoders (we use cross-encoder/ms-marco-MiniLM-L-6-v2 by default.) on the query-passage pairs that we have so far (for both positive and negative examples).
Result file (under path
$path_to_generated_data):gpl-training-data.tsv. It contains (gpl_steps*batch_size_gpl) tuples in total.
Up to now, we have the actual training data ready. One can look at sample-data/generated/fiqa for a quick example about the data format. The very last step is to apply the MarginMSE loss to teach the student retriever to mimic the margin scores, CE(query, positive) - CE(query, negative) labeled by the teacher model (Cross-Encoder, CE). And of course, the MarginMSE step is included in GPL and will be done automatically:). Note that MarginMSE works with dot-product and thus the final models trained with GPL works with dot-product.
PS: The --retrievers are for negative mining. They can be any dense retrievers trained on the general domain (e.g. MS MARCO) and do not need to be strong for the target task/domain. Please refer to the paper for more details (cf. Table 7).
One can also replace/put the customized data for any intermediate step under the path $path_to_generated_data with the same name fashion. GPL will skip the intermediate steps by using these provided data.
As a typical workflow, one might only have the (English) unlabeld corpus and want a good model performing well for this corpus. To run GPL training under such condition, one just needs these steps:
- Prepare your corpus in the same format as the data sample;
- Put your
corpus.jsonlunder a folder, e.g. named as "generated" for data loading and data generation by GPL; - Call gpl.train with the folder path as an input argument: (other arguments work as usual)
python -m gpl.train \
--path_to_generated_data "generated" \
--output_dir "output" \
--new_size -1 \
--queries_per_passage -1We now release the pre-trained GPL models via the https://huggingface.co/GPL. There are currently five types of models:
GPL/${dataset}-msmarco-distilbert-gpl: Model with training order of (1) MarginMSE on MSMARCO -> (2) GPL on${dataset};GPL/${dataset}-tsdae-msmarco-distilbert-gpl: Model with training order of (1) TSDAE on${dataset}-> (2) MarginMSE on MSMARCO -> (3) GPL on${dataset};GPL/msmarco-distilbert-margin-mse: Model trained on MSMARCO with MarginMSE;GPL/${dataset}-tsdae-msmarco-distilbert-margin-mse: Model with training order of (1) TSDAE on ${dataset} -> (2) MarginMSE on MSMARCO;GPL/${dataset}-distilbert-tas-b-gpl-self_miner: Starting from the tas-b model, the models were trained with GPL on the target corpus${dataset}with the base model itself as the negative miner (here noted as "self_miner").
Models 1. and 2. were actually trained on top of models 3. and 4. resp. All GPL models were trained the automatic setting of new_size and queries_per_passage (by setting them to -1). This automatic setting can keep the performance while being efficient. For more details, please refer to the section 4.1 in the paper.
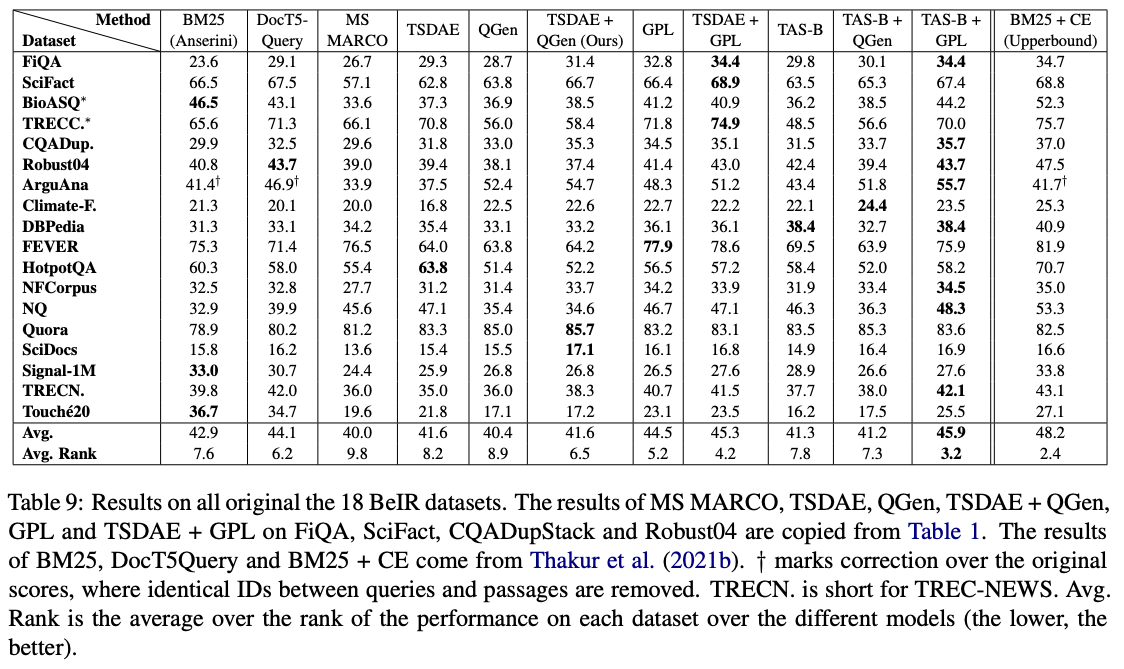
Among these models, GPL/${dataset}-distilbert-tas-b-gpl-self_miner ones works the best on the BeIR benchmark:
For reproducing the results with the same package versions used in the experiments, please refer to the conda environment file, environment.yml.
We now release the generated data used in the experiments of the GPL paper:
- The generated data for the main experiments on the 6 BeIR datasets: https://public.ukp.informatik.tu-darmstadt.de/kwang/gpl/generated-data/main/;
- The generated data for the experiments on the full 18 BeIR datasets: https://public.ukp.informatik.tu-darmstadt.de/kwang/gpl/generated-data/beir.
Please note that the 4 datasets of bioasq, robust04, trec-news and signal1m are only available after registration with the original official authorities. We only release the document IDs for these corpora with the file name corpus.doc_ids.txt. For more details, please refer to the BeIR repository.
If you use the code for evaluation, feel free to cite our publication GPL: Generative Pseudo Labeling for Unsupervised Domain Adaptation of Dense Retrieval:
@article{wang2021gpl,
title = "GPL: Generative Pseudo Labeling for Unsupervised Domain Adaptation of Dense Retrieval",
author = "Kexin Wang and Nandan Thakur and Nils Reimers and Iryna Gurevych",
journal= "arXiv preprint arXiv:2112.07577",
month = "4",
year = "2021",
url = "https://arxiv.org/abs/2112.07577",
}Contact person and main contributor: Kexin Wang, kexin.wang.2049@gmail.com
https://www.ukp.tu-darmstadt.de/
Don't hesitate to send us an e-mail or report an issue, if something is broken (and it shouldn't be) or if you have further questions.
This repository contains experimental software and is published for the sole purpose of giving additional background details on the respective publication.