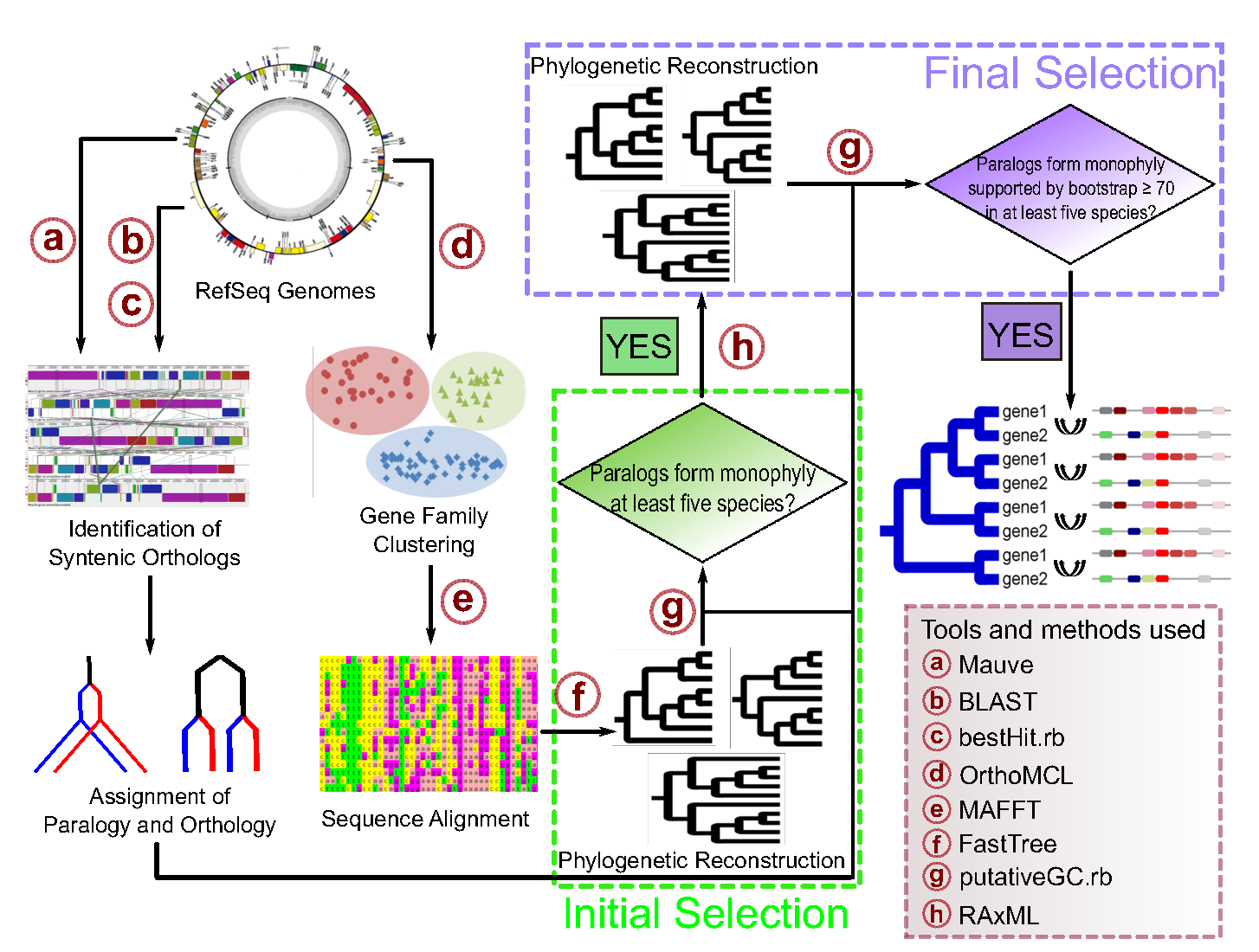
iSeeCE is a bioinformatic pipeline that allows identifying recurrent gene conversion across lineages.
v1.1 (2020.09.19)
- some bugs related to running OrthoMCL fixed thanks to the help of alexandergz1983. Now it is required to provide the configuration file for OrthoMCL when using "iSeeCE.sh".
- diamond instead of blastp allowed by specifying "--diamond"
iSeeCE is run in Linux environment.
Many scripts included in this product require packages from
This product also employs several other computational tools. Please ensure that you have them installed.
Important: Please be aware that it is important to change "dbLogin" and "dbPassword" in the configuration file of OrthoMCL. An example file is provided with iSeeCE (orthomcl.config)).
To see whether required software and libraries have been installed, please type
bash check_requirements.shiSeeCE identifies recurrent concerted evolution (CE) or gene conversion (GC) across species based on gene phylogeny and synteny. In brief, paralogs from the same species that form monophyly and have syntenic (positional) orthologs across species will be identified as converted genes. For visualiztion of some examples, please see our paper at the end or visit our database at www.lrgcdb.eu.
The input files should be genomes in the format of Genbank. Please make sure that the nucleotide sequences of the genome are included in each Genbank-formatted input file.
For the full usage, please type
bash iSeeCE.sh -hAs follows are some examples:
# default: gc_count_min=2, bootstrap=0.5
bash iseeCE.sh --indir genebank_dir --outdir new_dir --orthomcl_config orthomcl_config_gile
# No. of >=8 converted duplicate gene pairs in the gene family phylogeny, each conversion event supported by bootstrap >= 0.9
bash iseeCE.sh --indir genebank_dir --outdir new_dir --orthomcl_config orthomcl_config_gile --gc_count_min 8 --bootstrap 0.9
# Use diamond (which accelerates all-against-all homolog search for OrthoMCL by >1000 times but with potential cost of lower sensitivity)
bash iseeCE.sh --indir genebank_dir --outdir new_dir --orthomcl_config orthomcl_config_gile --diamond
# Ten threads for BLAST and MAFFT
bash iseeCE.sh --indir genebank_dir --outdir new_dir --orthomcl_config orthomcl_config_gile --cpu 10
# Five threads for BLAST and three threads for MAFFT
bash iseeCE.sh --indir genebank_dir --outdir new_dir --orthomcl_config orthomcl_config_gile --blast_cpu 5 --mafft_cpu 3
By default, syntenic orthologs will be identified by using only best reciprocal BLAST hits. The results of converted genes will be output to the file FastTree/GC.result.
Sometimes, concerted evolution or gene conversion can be difficult to be distinguished from tandem duplication. Thus, tandem duplicates will be identified and output to TD/TD.list. Clusters of tandem duplicates are defined as paralogs separated by no more than five genes and located within 20kb on the chromosome. Tandem duplicates are those from the same tandem duplicate clusters. All tandem duplicates are labeled with a star in the output FastTree/GC.result.
When "--mauve" is specified, syntenic orthologs will additionally be identified by Mauve whose results will be output to the file FastTree/GC.mauve.result. Also, it is necessary to specify the path to Mauve.jar and progressiveMauve using --mauve_jar and --mauve_prog, respectively. So please make sure that Mauve.jar has been installed if you want to perform the analysis with Mauve. Note that '--mauve' is optional. When '--mauve' is NOT specified, the analysis will be performed based on the best reciprocal BLAST hits. Important: '--mauve' is not recommended for analysis involving bacteria from different genera or even more distantly related strains as Mauve may be conservative (in other words, you will find very few converted genes in such situations), but it is always good to compare the result generated by "--mauve" with that not using "--mauve".
When '--bootstrap' is specified, only paralogs with support value equal to or above the given value will be analyzed. However, it should be noted that here 'bootstrap' refers to the support value obtained by Shimodaira-Hasegawa test implemented with FastTree, rather than regular bootstrap value. For more details, please visit http://www.microbesonline.org/fasttree/.
Please see CC BY 4.0 for licensing information.
You are welcome to cite our paper (see below) for iSeeCE if you feel it is useful.
You may need to cite corresponding papers for the use of OrthoMCL, diamond, MAFFT, FastTree, RAxML, Mauve, BioRuby and BioPerl.
Wang, S., & Chen, Y. (2018). Phylogenomic analysis demonstrates a pattern of rare and long-lasting concerted evolution in prokaryotes. Communications biology, 1(1), 1-11.
We thank Michael Imelfort and Kranti Konganti for sharing the free script gff2fasta.pl for sequence processing.