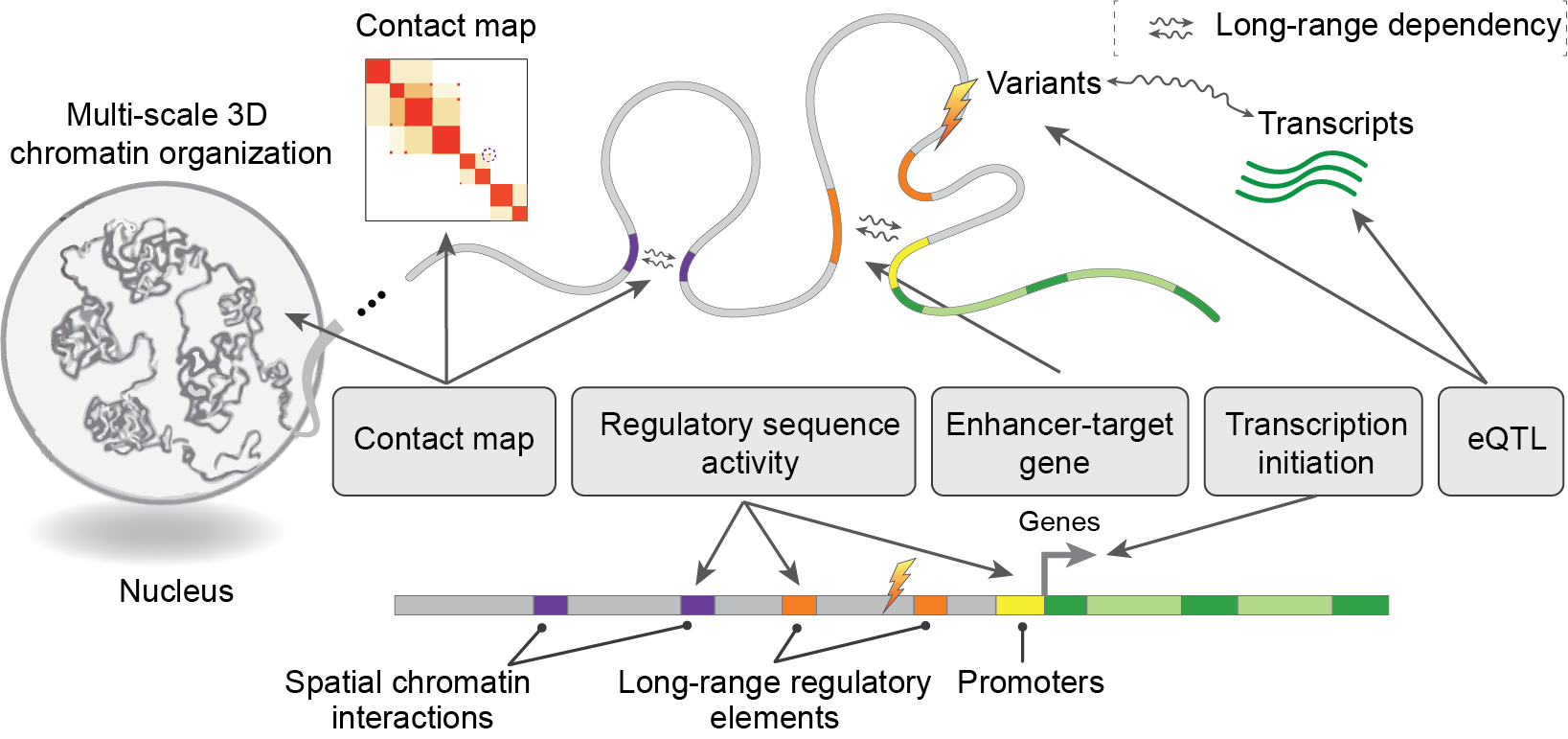
DNALongBench is a benchmark of realistic and biologically meaningful genomic DNA prediction tasks that require long-range sequence input and involve long-range dependencies. There are five tasks in our DNALongBench.
| LR Tasks | LR Type | Input Length | Output Shape | # Samples | Metric |
|---|---|---|---|---|---|
| Enhancer-target Gene | Binary Classification | 450,000 | 1 | 2,602 | AUROC |
| eQTL | Binary Classification | 450,000 | 1 | 31,282 | AUROC |
| Contact Map | Binned (2048bp) 2D Regression | 1,048,576 | 99,681 | 7,840 | SCC & PCC |
| Regulatory Sequence Activity | Binned (128bp) 1D Regression | 196,608 | Human: (896, 5,313) Mouse: (896, 1,643) |
Human: 38,171 Mouse: 33,521 |
PCC |
| Transcription Initiation Signal | Nucleotide-wise 1D Regression | 100,000 | (100,000, 10) | 100,000* | PCC |
The data for each task could be downloaded via the following link. Alternatively, you could download the data from Box.
The data can be downloaded at Regulatory Sequence Activity Prediction.
We provide the sequences.bed, statistics.json, hg38.ml.fa.fai and hg38.ml.fa.gz files, and reformulate these data into train*/valid*/test*.tfr. The only files you needed are the corresponding tfr files.
The data can be downloaded at Transcription Initiation Signal Prediction.
We provide all the correspoding bed files.
The data can be downloaded at Enhancer-Target Gene Prediction.
The sequences, fa and metrics data are provided.
The data can be downloaded at Contact Map Prediction.
We provide the well-split train/valid/test files.
The data can be downloaded at eQTL.
The corresponding bed files are provided.
We've provided the performance of three types of models, which are Expert Model, a lightweight CNN baseline, and a finetuned DNA foundation model (HyenaDNA, Caduceus-Ph and Caduceus-PS). We'll introduce below how to run these models by taking the task of Enhancer-Target Gene Prediction (ETGP) as an example.
| Model | Expert Model | CNN | HyenaDNA | Caduceus-Ph | Caduceus-PS |
|---|---|---|---|---|---|
| ETGP | 0.926 | 0.797 | 0.828 | 0.826 | 0.821 |
Following the commands below to download our code:
conda create -n dnalongbench python=3.9 -y
conda activate dnalongbench
git clone https://github.com/wenduocheng/DNALongBench.git
pip install .import dnalongbench
from dnalongbench.utils import load_data
train_loader, valid_loader, test_loader = load_data(root=root, task_name = 'contact_map_prediction', subset='HFF', batch_size=16)We also provide data loaders for each task in scripts/data_loaders.ipynb.
Please refer to experiments/CNN/README.md.
We used the official code of HyenaDNA. The environment setup can be found at HyenaDNA Enviroment Setup.
Be careful if you would like to use flash attention. Sometimes there are some issues when installing flash attention. We recommend first setup the environment, then activate the enviroment, and finally install flash attention inside the environment.
For RSAP and TISP tasks, please refer to the readme under experiments/HyenaDNA_RSAP_TISP/README.md
experiments/HyenaDNA/HyenaDNA_RSAP_TISP/README.mdFor CMP, eQTLP and ETGP tasks, please refer to experiments/HyenaDNA_ETGP_CMP_eQTLP/README.md
experiments/HyenaDNA/HyenaDNA_ETGP_CMP_eQTLP/README.mdWe used the official environment provided by Caduceus.
To get started, create a conda environment containing the required dependencies.
cd experiments/Caduceus/Caduceus_CMP_eQTLP_ETGP
conda env create -f caduceus_env.ymlActivate the environment.
conda activate caduceus_envFor RSAP and TISP tasks, please refer to experiments/Caduceus_RSAP_TISP/README.md
experiments/Caduceus_RSAP_TISP/README.mdFor CMP, eQTLP and ETGP tasks, please refer to experiments/Caduceus_CMP_eQTLP_ETGP/README.md
experiments/Caduceus/Caduceus_CMP_eQTLP_ETGP/README.mdWe used the official environment setup of GENERator: https://github.com/GenerTeam/GENERator.git.
git clone https://github.com/GenerTeam/GENERator.git
cd GENERator
pip install -r requirements.txtPlease refer to experiments/GENERator/README.md.
experiments/GENERator/README.mdWe used the official environment setup of Evo2.
Evo 2 is based on StripedHyena 2 which requires python>=3.11. Evo 2 uses Transformer Engine FP8 for some layers which requires an H100 (or other GPU with compute capability ≥8.9). We are actively investigating ways to avoid this requirement.
To install Evo 2 for inference or generation, please clone and install from GitHub. We recommend using a new conda environment with python>=3.11.
git clone --recurse-submodules git@github.com:ArcInstitute/evo2.git
cd evo2
pip install .Please refer to experiments/Evo2/README.md.
experiments/Evo2/README.md@inproceedings{chengdna,
title={DNALongBench: A Benchmark Suite for Long-Range DNA Prediction Tasks},
author={Cheng, Wenduo and Song, Zhenqiao and Zhang, Yang and Wang, Shike and Wang, Danqing and Yang, Muyu and Li, Lei and Ma, Jian}
}