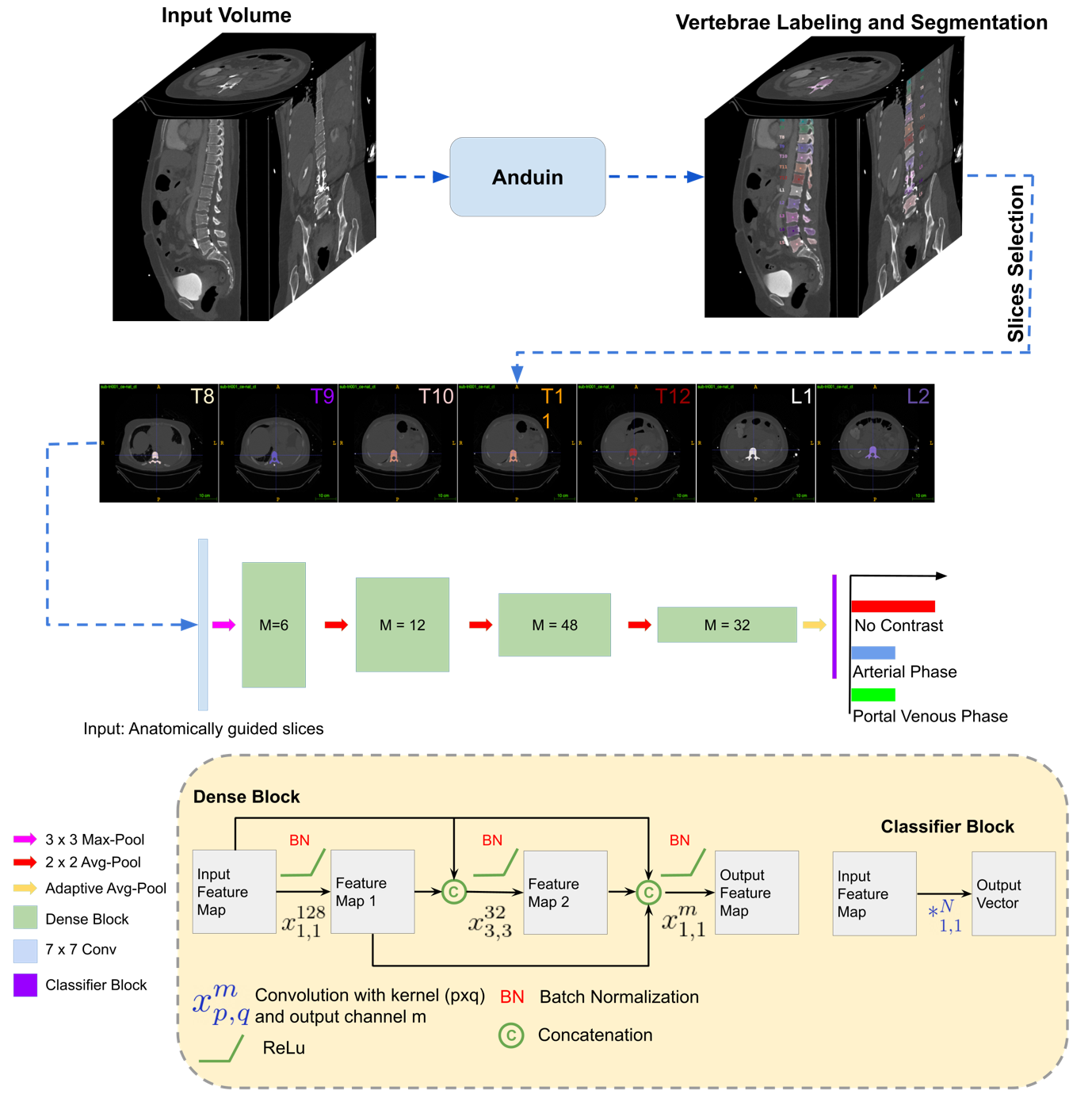
#Automated detection of the contrast phase in MDCT by an artificial neural network improves accuracy of opportunistic bone mineral density measurements
Source code for our paper "Automated detection of the contrast phase in MDCT by an artificial neural network improves accuracy of opportunistic bone mineral density measurements" as described in Jounal of Europena Radiology.
Authors: Sebastian Rühling, Fernando Navarro,
You need to have following in order for this library to work as expected
- python >= 3.6.5
- pip >= 18.1
- tensorflow-gpu = 1.9.0
- tensofboard = 1.9.0
- numpy >= 1.15.0
- dipy >= 0.14.0
- matplotlib>= 2.2.2
- nibabel >= 1.15.0
- pandas >= 0.23.4
- scikit-image >= 0.14.0
- scikit-learn >= 0.20.0
- scipy >= 1.1.0
- seaborn >= 0.9.0
- SimpleITK >= 1.1.0
- tabulate >= 0.8.2
- xlrd >= 1.1.0
Run pip install -r requirements.txt
-
Download VerSe19 and Verse20 for sample data VerSe19 VerSe20.
-
Obtain Vertebrae Segmentations using Anduin.
-
Your data must be in the following structure:
└── data_folder
├── raw_data
├── patient001
├── patient001_ct_image.nii.gz
├── derivatives
├── patient001
├── patient001_seg-subreg_ctd.json
- Convert your data into npz files before starting the training using the command:
python data2npz.py --data_folder ./data --dst_dolder ./npz_files
Script argments:
--data_folder ./data Path to the model to the data folder containing NIfTI files(default: ./data)
--task {play,eval,train}
task to perform, must load a pretrained model if task
is "play" or "eval" (default: train)
--file_type {brain,cardiac,fetal}
Type of the training and validation files (default:
train)
--files FILES [FILES ...]
Filepath to the text file that contains list of
images. Each line of this file is a full path to an
image scan. For (task == train or eval) there should
be two input files ['images', 'landmarks'] (default:
None)
--val_files VAL_FILES [VAL_FILES ...]
Filepath to the text file that contains list of
validation images. Each line of this file is a full
path to an image scan. For (task == train or eval)
there should be two input files ['images',
'landmarks'] (default: None)
--saveGif Save gif image of the game (default: False)
--saveVideo Save video of the game (default: False)
--logDir LOGDIR Store logs in this directory during training (default:
runs)
--landmarks [LANDMARKS [LANDMARKS ...]]
Landmarks to use in the images (default: [1])
--model_name {CommNet,Network3d}
Models implemented are: Network3d, CommNet (default:
CommNet)
--batch_size BATCH_SIZE
Size of each batch (default: 64)
--memory_size MEMORY_SIZE
Number of transitions stored in exp replay buffer. If
too much is allocated training may abruptly stop.
(default: 100000.0)
--init_memory_size INIT_MEMORY_SIZE
Number of transitions stored in exp replay before
training (default: 30000.0)
--max_episodes MAX_EPISODES
"Number of episodes to train for" (default: 100000.0)
--steps_per_episode STEPS_PER_EPISODE
Maximum steps per episode (default: 200)
--target_update_freq TARGET_UPDATE_FREQ
Number of epochs between each target network update
(default: 10)
--save_freq SAVE_FREQ
Saves network every save_freq steps (default: 1000)
--delta DELTA Amount to decreases epsilon each episode, for the
epsilon-greedy policy (default: 0.0001)
--viz VIZ Size of the window, None for no visualisation
(default: 0.01)
--multiscale Reduces size of voxel around the agent when it
oscillates (default: False)
--write Saves the training logs (default: False)
--train_freq TRAIN_FREQ
Number of agent steps between each training step on
one mini-batch (default: 1)
--seed SEED Random seed for both training and evaluating. If none
is provided, no seed will be set (default: None)
- Run the training script with the specified parameters:
python train_anatomy_guided.py --data_folder ./data --dst_dolder ./npz_files
Run the command:
python infer_anatomy_guided.py --data_folder ./data --dst_dolder ./npz_files
Please cite our paper if it is useful for your research:
@article{navarro2019shape,
title={Shape-Aware Complementary-Task Learning for Multi-Organ Segmentation},
author={Navarro, Fernando and Shit, Suprosanna and Ezhov, Ivan and Paetzold, Johannes and Gafita, Andrei and Peeken, Jan and Combs, Stephanie and Menze, Bjoern},
journal={arXiv preprint arXiv:1908.05099},
year={2019}
}
- Fernando Navarro - ferchonavarro
Let us know if you face any issues. You are always welcome to report new issues and bugs and also suggest further improvements. And if you like our work hit that start button on top. Enjoy :)