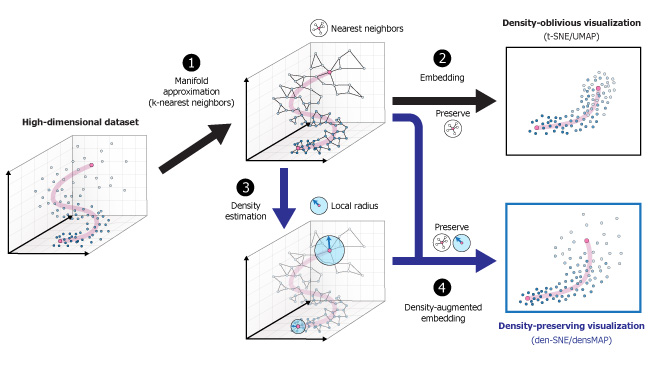
This repository contains density-preserving data visualization tools den-SNE and densMAP, augmenting the tools t-SNE and UMAP respectively.
A detailed README for each algorithm (including installation notes, usage guidelines, and example scripts) is provided in its respective subdirectory. Direct links: den-SNE and densMAP.
Our algorithms are described in the following manuscript:
Ashwin Narayan, Bonnie Berger*, and Hyunghoon Cho*, "Assessing single-cell transcriptomic variability through density-preserving data visualization", Nature Biotechnology, 2021
and an earlier preprint:
Ashwin Narayan, Bonnie Berger*, and Hyunghoon Cho*. "Density-Preserving Data Visualization Unveils Dynamic Patterns of Single-Cell Transcriptomic Variability", bioRxiv, 2020.
densMAP is now available as part of the Python UMAP package.
Our den-SNE software is based on the Barnes-Hut implementation of t-SNE (https://github.com/lvdmaaten/bhtsne) described in the following paper:
Laurens Van Der Maaten. "Accelerating t-SNE using Tree-based Algorithms", Journal of Machine Learning Research, 15(1), 2014.
Our densMAP software is based on the implementation of UMAP (https://github.com/lmcinnes/umap) described in the following paper:
Leland McInnes, John Healy, and James Melville. "UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction", arXiv, 1802.03426, 2018.
Both den-SNE and densMAP are licensed under the MIT license. See each subdirectory for detail.
Ashwin Narayan, ashwinn@mit.edu
Hyunghoon Cho, hhcho@broadinstitute.org