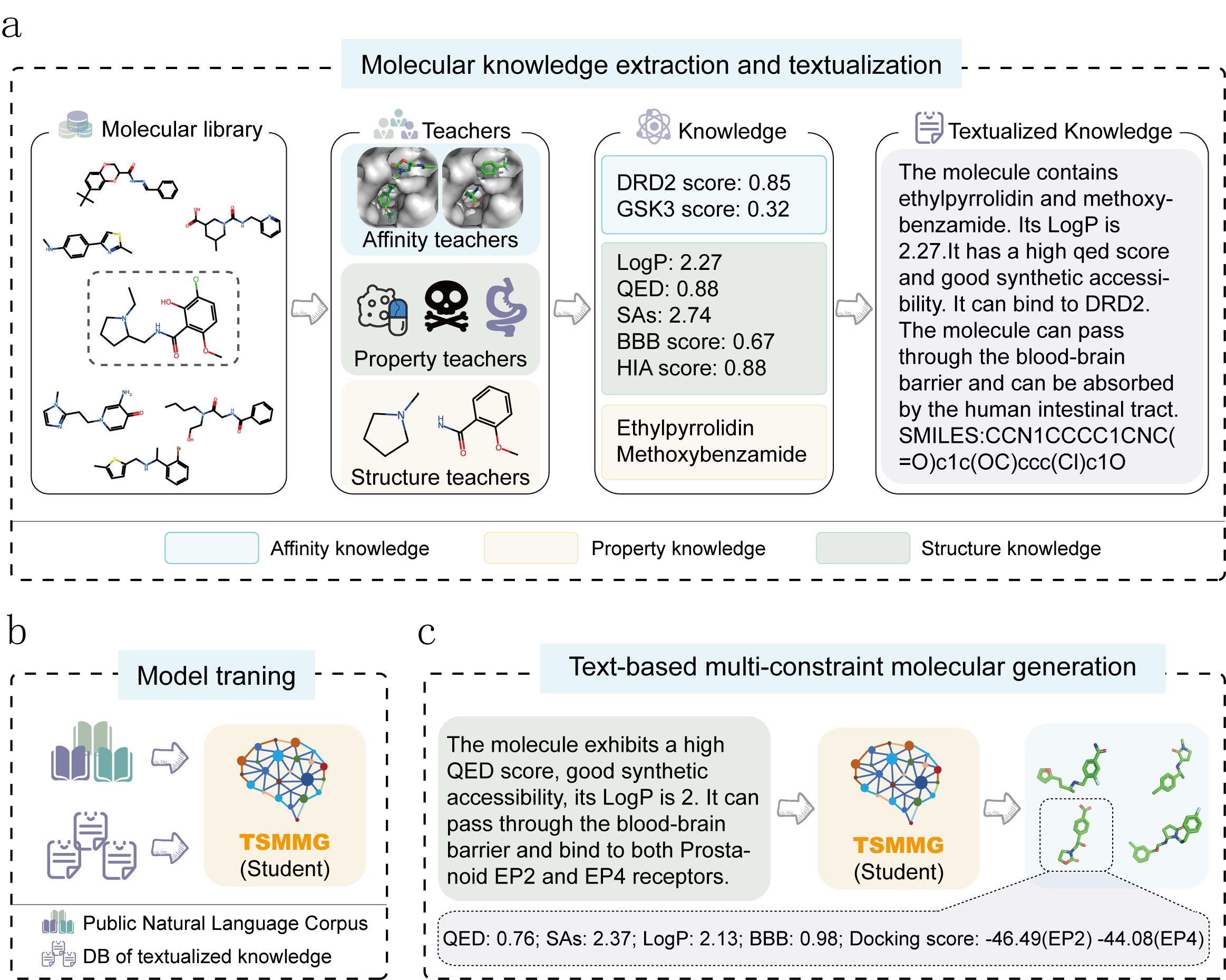
Instruction Multi-Constraint Molecular Generation Using a Teacher-Student Large Language Model
While various models and computational tools have been proposed for structure and property analysis of molecules, generating molecules that conform to all desired structures and properties remains a challenge. Here, we introduce a multi-constraint molecular generation large language model, TSMMG, which, akin to a student, incorporates knowledge from various small models and tools, namely, the 'teachers'. To train TSMMG, we construct a large set of text-molecule pairs by extracting molecular knowledge from these 'teachers', enabling it to generate novel molecules that conform to the descriptions through various text prompts. We experimentally show that TSMMG remarkably performs in generating molecules meeting complex, natural language-described property requirements across two-, three-, and four-constraint tasks, with an average molecular validity of over 99% and success ratio of 88.08%, 65.27%, and 61.44%, respectively. The model also exhibits adaptability through zero-shot testing, creating molecules that satisfy combinations of properties that have not been encountered. It can comprehend text inputs with various language styles, extending beyond the confines of outlined prompts, as confirmed through empirical validation. Additionally, the knowledge distillation feature of TSMMG contributes to the continuous enhancement of small models, while the innovative approach to dataset construction effectively addresses the issues of data scarcity and quality, which positions TSMMG as a promising tool in the domains of drug discovery and materials science.
conda create -n tsmmg python=3.10.9
conda activate tsmmg
pip install torch==2.0.1 torchvision==0.15.2 torchaudio==2.0.2 # cuda 11.7
pip install transformers
pip install rdkit
pip install pandas
pip install accelerate
pip install weasyprint
pip install libsvm-official
pip install matplotlib
pip install datasetsDownload the necessary data for this project from either Baidu Cloud or Google Drive. Please choose the platform that is most convenient for you.
Move the downloaded TSMMG.zip file to the same directory as the TSMMG source code. For example:
pwd
>> /home/gary
ls
>> TSMMG TSMMG.zipNext, unzip the compressed data files:
unzip TSMMG.zip
cd TSMMG/model_weights
tar -zxvf smiles2iupac.tar.gz # this is the smiles2iupac model for evaluation
cd checkpoints/gpt2_ft
tar -zxvf checkpoint-1360000.tar.gz # this is an example of trained weightsFinally, unzip the compressed source files:
cd /home/gary/TSMMG
tar -zxvf TARGET.tar.gz
tar -zxvf ADMET.tar.gzTo train the model, use the following command:
accelerate launch train.py --backbone=gpt2 --learning_rate=5e-4 --training_strategy=ft --epochs=20 --quantify=no --batch_size=32We've provided an example of trained weights, so you can directly run the following code for evaluation without needing to train a new model.
Use the following command for evaluation:
CUDA_VISIBLE_DEVICES=[CUDA_NO] python eval_v2.py --backbone=gpt2 --training_strategy=ft --eval_type=[TASK_NAME] --checkpoint=1360000 --return_num=5 --quantify=no --T=1In the above commands, replace [CUDA_NO] with the number of the CUDA device you want to use, [TASK_NAME] with the task you want to evaluate.
For examples:
CUDA_VISIBLE_DEVICES=0 python eval_v2.py --backbone=gpt2 --training_strategy=ft --eval_type=drd2 --checkpoint=1360000 --return_num=5 --quantify=no --T=1The inference time will be approximately 1 minute for 500 molecules (on Tesla V100).
The generated SMILES will be in the file './outputs/ouputs_drd2.csv'.
For more information on the supported tasks, please refer to the 'TSMMG/data/eval/' directory.
cd /home/gary/TSMMG/metric
python run_metric.py --backbone=gpt2 --training_strategy=ft --checkpoint=1360000 --eval_type=drd2The evaluation results will be in the file '/home/gary/TSMMG/metric/outputs/output_gpt2_ft_drd2_mt.csv' and '/home/gary/TSMMG/metric/result/gpt2_ft_1360000_drd2-IsValid,DRD2.csv'
- Xiangxiang Zeng, Peng Zhou, Jianmin Wang et al. Instruction Multi-Constraint Molecular Generation Using a Teacher-Student Large Language Model, 19 March 2024, PREPRINT (Version 1) available at Research Square [https://doi.org/10.21203/rs.3.rs-3845824/v1]