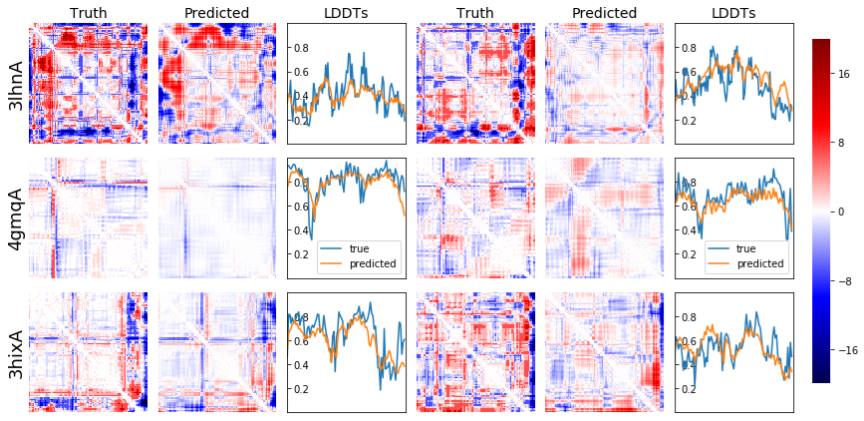
A script for predicting protein model accuracy.
usage: ErrorPredictor.py [-h] [--pdb] [--multiDecoy] [--reference]
[--noEnsemble] [--leavetemp] [--verbose]
[--process PROCESS] [--gpu GPU] [--featurize]
[--reprocess]
infolder ...
Error predictor network
positional arguments:
infolder input folder name full of pdbs or path to a single pdb
outfolder output folder name. If a pdb path is passed this needs
to be a .npz file. Can also be empty. Default is
current folder or pdbname.npz
optional arguments:
-h, --help show this help message and exit
--pdb, -pdb Running on a single pdb file instead of a folder
(Default: False)
--multiDecoy, -mm running the multi-model option (Default: False)
--reference, -ref running the reference model trained based on distance
information only. (Default: False)
--noEnsemble, -ne running without model ensembling (Default: False)
--leavetemp, -lt leaving temporary files (Default: False)
--verbose, -v verbose flag (Default: False)
--process PROCESS, -p PROCESS
# of cpus to use for featurization (Default: 1)
--gpu GPU, -g GPU gpu device to use (default gpu0)
--featurize, -f running only featurization (Default: False)
--reprocess, -r reprocessing all feature files (Default: False)
v0.0.1
Type the following commands to activate tensorflow environment with pyrosetta3.
source activate tensorflow
source /software/pyrosetta3/setup.sh
Running on a folder of pdbs (foldername: samples)
python ErrorPredictor.py -r -v samples outputs
Running on a single pdb file (inputname: input.pdb). Output name is optional and defaults to input.npz
python ErrorPredictor.py -r -v --pdb input.pdb [output.npz]
Only doing the feature processing (foldername: samples)
python ErrorPredictor.py -r -v -f samples outputs
Output of the network is written to [input_file_name].npz.
You can extract the predictions as follows.
import numpy as np
x = np.load("testoutput.npz")
lddt = x["lddt"] # per residue lddt
estogram = x["estogram"] # per pairwise distance e-stogram
mask = x["mask"] # mask predicting native < 15
Perhaps lddt is the easiest place to start as it is per-residue quality score. You can simply take an average if you want a global score per protein structure.
If you want to do something more involved, especially for protein complex design, see example.ipynb for getting more specialized metrics. If you want to play with pair-wise error predictions, samples.ipynb is a good place to start.
- If ErrorPredictor.py returns an OOM (out of memory) error, your protein is probably too big. Try getting on titan instead of rtx2080 or run without gpu if running time is not your problem. You can also truncate your protein structures although it is not recommended.
- If you get an import error for pyErrorPred, you probably moved the script out of LocalAccuacyPredictor. In that case, you would have to add pyErrorPred to python path or do so within the script.
- Send an e-mail at hiranumn at cs dot washington dot edu.
- Python3.5>
- Pyrosetta
- Tensorflow 1.14 (not Tensorflow 2.0)
- Added reference state mode, 2019.12.4
- Reorganized code so that it is a python package, 2019.11.10
- Added some analysis code, 2019.11.6
- Distance matrix calculation speed-up, 2019.10.25
- v 0.0.1 released, 2019.10.19