A minimalist roadmap to the Data Science World, based on the Kaggle's courses, plus some COOL stuffs. For the Kaggle's courses, all credits goes to their instructors.
Python
Pandas
Geospatial Analysis
Intro to Machine Learning
Intermediate Machine Learning
Deep Learning
Intro to SQL
Machine Learning Explainability
Learn the most important language for data science.
A quick introduction to Python syntax, variable assignment, and numbers. #
=func(var)
var.func()/ # true division
// # floor division
% # modulus
** # exponentiationmin()
max()
abs()# conversion functions
int()
float()Calling functions and defining our own, and using Python's builtin documentation. #
on modules, objects, instances, and ...
help()
dir()def func_name(vars):
# some useful codes
return # some useful results""" some useful info about the function """ # `help()` returns thisprint()print(..., sep="\t")fn(fn(arg))
string.lower().split()Using booleans for branching logic. #
True
False
bool()a == b # a equal to b
a != b # a not equal to b
a < b # a less than b
a > b # a greater than b
a <= b # a less than or equal to b
a >= b # a greater than or equal to bPEMDAS combined with Boolean Values
()
**
+x, -x, ~x
*, /, //, %
+, -
<<, >>
&
^
|
==, !=, >, >=, <, <=, is, is not, in, not in
not
and
orif
elif
else- All numbers are treated as
True, except0. - All strings are treated as
True, except the empty string"". - Empty sequences (strings, lists, tuples, sets) are
Falseand the rest areTrue.
Setting a variable to either of two values depending on a condition.
outcome = "failed" if grade < 50 else "passed"Lists and the things you can do with them. Includes indexing, slicing and mutating. #
A mutable mix of same or different types of variables
[]
list()planets = ["Mercury", "Venus", "Earth", "Mars", "Jupiter"]
planets[0] # first element
planets[-1] # last elementplanets[:3]
planets[-3:]planets[:3] = ["Mur", "Vee", "Ur"]len()
sorted()
max()
sum()
any()Everything is an Object.
# complex number object
c = 12 + 5j
c.imag
c.real# integer number object
x = 12
x.bit_length()list.append()
list.pop()
list.index()
inImmutable.
()
,
tuple()x = 0.125
numerator, denominator = x.as_integer_ratio()For and while loops, and a much-loved Python feature: list comprehensions. #
Use in every iteratable objects: list, tuples, strings, ...
for - in - :
# some useful codesrange()while
[- for - in -]squares = [n ** 2 for n in range(10)]
# constant
[32 for planet in planets]# with if
short_planets = [planet.upper() + "!" for planet in planets if len(planet) < 6]# combined with other functions
return len([num for num in nums if num < 0])
return sum([num < 0 for num in nums])
return any([num % 7 == 0 for num in nums])Solving a problem with less code is always nice, but it's worth keeping in mind the following lines from The Zen of Python.
Readability counts.
Explicit is better than implicit.
for index, item in enumerate(items):
# some useful codesWorking with strings and dictionaries, two fundamental Python data types. #
Immutable.
""
""" """
str()[char + "! " for char in "Planet"]
>>> ['P! ', 'l! ', 'a! ', 'n! ', 'e! ', 't! ']"Planet"[0] = "M"
>>> TypeError: 'str' object does not support item assignmentstr.upper()
str.lower()
str.index()
str.startswith()
str.endswith()# split
year, month, day = '2020-03-05'.split('-')
year, month, day
>>> ('2020', '03', '05')# join
'/'.join([month, day, year])
>>> '03/05/2020'"{}".format()
f"{}"Pairs of keys,values.
{}
dict()numbers = {"one": 1, "two": 2, "three": 3}
numbers["one"]
numbers["eleven"] = 11planet_to_initial = {planet: planet[0] for planet in planets}dict.keys()
dict.values()" ".join(sorted(planet_to_initial.values()))key_of_min_value = min(numbers, key=numbers.get)"M" in planet_to_initial.values()
>>> True# loop over keys
for planet in planet_to_initial:
print(planet)# loop over (key, value) pairs using `item`
for planet, initial in planet_to_initial.items():
print(f'{planet} begins with "{initial}"')Imports, operator overloading, and survival tips for venturing into the world of external libraries. #
# simple import, `.` access
import math
math.pi# `as` import, short `.` access
import math as mt
mt.pi# `*` import, simple access
from math import *
piThe problem of * import is that some modules (ex. math and numpy) have functions with same name (ex. log) but with different semantics. So one of them overwrites (or "shadows") the other. It is called overloading.
# combined, solution for the `*` import
from math import log, pi
from numpy import asarrayModules contain variables which can refer to functions or values. Sometimes they can also have variables referring to other modules.
import numpy
dir(numpy.random)
>>> ['set_state', 'shuffle', 'standard_cauchy', 'standard_exponential', 'standard_gamma', 'standard_normal', 'standard_t', 'test', 'triangular', 'uniform', ...]# make an array of random numbers
rolls = numpy.random.randint(low=1, high=6, size=10)
rolls
>>> array([3, 2, 5, 2, 4, 2, 2, 3, 2, 3])Standard Python datatypes are: int, float, bool, list, str, and dict.
As you work with various libraries for specialized tasks, you'll find that they define their own types. For example
- Matplotlib:
Subplot,Figure,TickMark, andAnnotation - Pandas:
DataFrameandSeries - Tensorflow:
Tensor
Use type() to find the type of an object. Use dir() and help() for more details.
dir(numpy.ndarray)
>>> [...,'__bool__', ..., '__delattr__', '__delitem__', '__dir__', ..., '__sizeof__', ..., 'max', 'mean', 'min', ..., 'sort', ..., 'sum', ..., 'tobytes', 'tofile', 'tolist', 'tostring', ...]# list
xlist = [[1, 2, 3], [2, 4, 6]]
xlist[1, -1]
>>> TypeError: list indices must be integers or slices, not tuple# numpy array
xarray = numpy.asarray(xlist)
xarray[1, -1]
>>> 6# list
[3, 4, 1, 2, 2, 1] + 10
>>> TypeError: can only concatenate list (not "int") to list# numpy array
rolls + 10
>>> array([13, 12, 15, 12, 14, 12, 12, 13, 12, 13])# tensorflow
import tensorflow as tf
a = tf.constant(1)
b = tf.constant(1)
a + b
>>> <tf.Tensor 'add:0' shape=() dtype=int32>When Python programmers want to define how operators behave on their types, they do so by implementing Dunder (Special) Methods, methods with special names beginning and ending with 2 underscores such as __add__ or __contains__. More info: https://is.gd/3zuhhL
The fundamental package for scientific computing with Python. #
NumPy’s main object is the homogeneous n-dimensional array. It is a table of elements (usually numbers), all of the same type, indexed by a tuple of non-negative integers. In NumPy dimensions are called axes. NumPy’s array class is called ndarray. Note that numpy.array is not the same as the Standard Python Library class array.array, which only handles one-dimensional arrays and offers less functionality.
# create an array with a single sequence as an argument
import numpy as np
a = np.array([2, 3, 4])
a.ndim # number of axes
>>> 1
a.shape # size of array in each dimension
>>> (1, 3)
a.size # total number of elements
>>> 3
a.dtype.name # type of elements
>>> 'int64'# sequences as arguments + dtype
c = np.array([[1, 2], [3, 4]], dtype=complex)
c
>>> array([[1.+0.j, 2.+0.j],
[3.+0.j, 4.+0.j]])NumPy offers several functions to create arrays with initial placeholder content. The function zeros creates an array full of zeros, the function ones creates an array full of ones, and the function empty creates an array whose initial content is random and depends on the state of the memory. By default, the dtype of the created array is float64.
np.ones((2, 3, 4), dtype=np.int16)
>>> array([[[1, 1, 1, 1],
[1, 1, 1, 1],
[1, 1, 1, 1]],
[[1, 1, 1, 1],
[1, 1, 1, 1],
[1, 1, 1, 1]]], dtype=int16)To create sequences of numbers, NumPy provides the arange function which is analogous to the Python built-in range, but returns an array.
a = np.arange(15).reshape(3, 5)
a
>>> array([[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14]])np.arange(0, 2, 0.3)
>>> array([0. , 0.3, 0.6, 0.9, 1.2, 1.5, 1.8])Function linspace receives as an argument the number of elements that we want, instead of the step. It's useful to evaluate function at lots of points
from numpy import pi
x = np.linspace(0, 2 * pi, 100)
f = np.sin(x)On NumPy arrays, arithmetic operators apply elementwise, even the product operator *, unlike in many matrix languages. The matrix product can be performed using the @ operator or the dot function. NumPy provides familiar mathematical functions such as sin, cos, and exp. Some operations, such as += and *=, act in place to modify an existing array rather than create a new one.
a = np.array([20, 30, 40, 50])
b = np.arange(4)
a - b
b ** 2
10 * np.sin(a)
a < 35A = np.array([[1, 1], [0, 1]])
B = np.array([[2, 0], [3, 4]])
A * B # elementwise product
A @ B # matrix product
A.dot(B) # another matrix productMany unary operations are implemented as methods of the ndarray class. By default, these operations apply to the array as though it were a list of numbers, regardless of its shape. However, by specifying the axis parameter we can apply an operation along the specified axis of an array.
# create an instance of a random generator
rg = np.random.default_rng(1)
a = rg.random((2, 3))
a.sum() # sum of array
a.min(axis=0) # min of each column
a.cumsum(axis=1) # cumulative sum along each rowHere is a list of some useful NumPy functions ordered in categories. See Routines for the full list.
-
Array Creation:
arange,array,copy,empty,empty_like,eye,fromfile,fromfunction,identity,linspace,logspace,mgrid,ogrid,ones,ones_like,r_,zeros,zeros_like -
Conversions:
astype,atleast_1d,atleast_2d,atleast_3d,mat -
Manipulations:
apply_along_axis,array_split,column_stack,concatenate,diagonal,dsplit,dstack,hsplit,hstack,item,newaxis,ravel,repeat,reshape,resize,squeeze,swapaxes,take,transpose,vsplit,vstack -
Questions:
all,any,nonzero,where -
Ordering:
argmax,argmin,argsort,max,min,ptp,searchsorted,sort -
Operations:
average,ceil,choose,compress,cumprod,conj,cumsum,diff,floor,inner,invert,fill,imag,prod,put,putmask,real,round,sum -
Basic Statistics:
cov,corrcoef,mean,median,std,var -
Basic Linear Algebra:
cross,dot,outer,linalg.svd,vdot
When operating with arrays of different types, the type of the resulting array corresponds to the more general or precise one (a behavior known as upcasting).
(np.ones(3, dtype='int8') + np.linspace(0, pi, 3)).dtype.name
>>> 'float64'One-dimensional arrays can be indexed, sliced and iterated over, much like lists and other Python sequences.
a = np.arange(10) ** 3 # the first 10 cube numbers
a[2:5]
a[:6:2] = 1000 # from start-to-6, set every 2nd element to 1000
a[::-1] # reversed aMultidimensional arrays can have one index per axis. These indices are given in a tuple.
def f(x, y):
return 10 * x + y
b = np.fromfunction(f, (5, 4), dtype=int)
b[2, 3]
b[:, 1] # each row in the second column of b
b[1:3, :] # each column in the second and third row of b
b[-1] # the last row. equivalent to b[-1,:]The expression within brackets in b[i] is treated as an i followed by as many instances of : as needed to represent the remaining axes. NumPy also allows you to write this using dots as b[i, ...]. The dots (...) represent as many colons as needed to produce a complete indexing tuple. For example, if x is an array with 5 axes, then x[1, 2, ...] is equivalent to x[1, 2, :, :, :].
Iterating over multidimensional arrays is done with respect to the first axis. However, if we want to perform an operation on each element in the array, we can use the flat attribute which is an iterator over all the elements of the array.
for element in b.flat:
print(element)NumPy offers more indexing facilities than regular Python sequences. In addition to indexing by integers and slices, as we saw before, arrays can be indexed by arrays of integers and arrays of booleans.
a = np.arange(12) ** 2 # the first 12 square numbers
i = np.array([1, 1, 3, 8, 5]) # an array of indices
a[i] # the elements of a at the positions i
>>> array([ 1, 1, 9, 64, 25])
j = np.array([[3, 4], [9, 7]]) # a bidimensional array of indices
a[j] # the same shape as j
>>> array([[ 9, 16],
[81, 49]])a = np.arange(12).reshape(3, 4)
a
>>> array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]])
i = np.array([[0, 1], [1, 2]])
j = np.array([[2, 1], [3, 3]])
a[i, j]
>>> array([[ 2, 5],
[ 7, 11]])
a[i, 2] # broadcasting
>>> array([[ 2, 6],
[ 6, 10]])We can also use indexing with arrays as a target to assign to.
a = np.arange(5)
a[[1, 3, 4]] = 0
a
>>> array([0, 0, 2, 0, 0])However, when the list of indices contains repetitions, the assignment is done several times, leaving behind the last value.
a = np.arange(5)
a[[0, 0, 2]] = [1, 2, 3]
a
>>> array([2, 1, 3, 3, 4])a = np.arange(5)
a[[0, 0, 2]] += 1
a
>>> array([1, 1, 3, 3, 4]) # the 0th element is only incremented onceWhen we index arrays with arrays of (integer) indices we are providing the list of indices to pick. With boolean indices the approach is different; we explicitly choose which items in the array we want and which ones we don’t. This can be very useful in assignments.
a = np.arange(12).reshape(3, 4)
b = a > 4
b # b is a boolean with a's shape
>>> array([[False, False, False, False],
[False, True, True, True],
[ True, True, True, True]])
a[b] # 1d array with the selected elements
>>> array([ 5, 6, 7, 8, 9, 10, 11])
a[b] = 0 # All elements of 'a' higher than 4 become 0
a
>>> array([[0, 1, 2, 3],
[4, 0, 0, 0],
[0, 0, 0, 0]])The second way of indexing with booleans is that for each dimension of the array we give a 1D boolean array selecting the slices we want.
a = np.arange(12).reshape(3, 4)
b1 = np.array([False, True, True]) # first dim selection
b2 = np.array([True, False, True, False]) # second dim selection
a[b1, :] # selecting rows
>>> array([[ 4, 5, 6, 7],
[ 8, 9, 10, 11]])
a[:, b2] # selecting columns
>>> array([[ 0, 2],
[ 4, 6],
[ 8, 10]])
a[b1][:, b2]
>>> array([[ 4, 6],
[ 8, 10]])
a[b1, b2] # a weird thing to do
>>> array([ 4, 10])The ix_ function can be used to construct an open mesh from multiple sequences. This function takes N 1-D sequences and returns N outputs with N dimensions each. Using ix_ one can quickly construct index arrays that will index the cross product.
a = np.arange(10).reshape(2, 5)
a
>>> array([[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9]])
ixgrid = np.ix_([0, 1], [2, 4])
ixgrid
>>> (array([[0],
[1]]), array([[2, 4]]))
ixgrid[0].shape, ixgrid[1].shape
>>> ((2, 1), (1, 2))
a[ixgrid]
>>> array([[2, 4],
[7, 9]])
ixgrid = np.ix_([True, True], [2, 4])
a[ixgrid]
>>> array([[2, 4],
[7, 9]])An array has a shape given by the number of elements along each axis. The shape of an array can be changed with various commands. Note that the following three commands all return a modified array, but do not change the original array.
a.ravel() # returns the array, flattened
a.transpose() # returns the array, transposed
a.reshape() # returns the array with a modified shapeThe reshape function returns its argument with a modified shape, whereas the resize function modifies the array itself. And, if a dimension is given as -1 in a reshaping operation, the other dimensions are automatically calculated.
Several arrays can be stacked together along different axes, vertically with vstack and horizontally with hstack. The function column_stack stacks 1D arrays as columns into a 2D array. It is equivalent to hstack only for 2D arrays. On the other hand, the function row_stack is equivalent to vstack for any input arrays. In general, for arrays with more than two dimensions, hstack stacks along their second axes, vstack stacks along their first axes, and concatenate allows for an optional arguments giving the number of the axis along which the concatenation should happen.
np.column_stack is np.hstack
>>> False
np.row_stack is np.vstack
>>> TrueUsing hsplit, we can split an array along its horizontal axis, either by specifying the number of equally shaped arrays to return, or by specifying the columns after which the division should occur. vsplit splits along the vertical axis, and array_split allows one to specify along which axis to split.
a = np.floor(10 * rg.random((2, 12)))
a
>>> array([[6., 7., 6., 9., 0., 5., 4., 0., 6., 8., 5., 2.],
[8., 5., 5., 7., 1., 8., 6., 7., 1., 8., 1., 0.]])
# split a into 3
np.hsplit(a, 3)
>>> [array([[6., 7., 6., 9.], [8., 5., 5., 7.]]),
array([[0., 5., 4., 0.], [1., 8., 6., 7.]]),
array([[6., 8., 5., 2.], [1., 8., 1., 0.]])]
# split a after the 3rd and the 4th column
np.hsplit(a, (3, 4))
>>> [array([[6., 7., 6.], [8., 5., 5.]]),
array([[9.], [7.]]),
array([[0., 5., 4., 0., 6., 8., 5., 2.], [1., 8., 6., 7., 1., 8., 1., 0.]])]When operating and manipulating arrays, their data is sometimes copied into a new array and sometimes not. There are three cases:
- No Copy at All - Simple assignments (
b = a) make no copy of objects or their data. - Shallow Copy or View - The
viewmethod creates a new array object that looks at the same data. Slicing an array returns a view of it too. Changing the data in a view, changes the base data. - Deep Copy - The
copymethod makes a complete copy of the array and its data. Sometimescopyshould be called after slicing if the original array, maybe a huge intermediate result, is not required anymore.
a = np.array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]])
b = a # no new object is created
b is a
>>> True
c = a[:, 1:3]
c.base is a
>>> True
c[:] = 10 # c[:] is a view of c, a's data changes
d = a.copy()
d.base is a # d doesn't share anything with a
>>> FalseSolve short hands-on challenges to perfect your data manipulation skills.
You can't work with data if you can't read it. #
It is a table and contains an array of individual entries. Each entry corresponds to a row (or record) and a column.
import pandas as pd
pd.DataFrame({"Apples": [50, 21], "Bananas": [131, 2]})The syntax for declaring a new one is a dictionary whose keys are the column names, and whose values are a list of entries.
The list of row labels used in a DataFrame is known as an Index. We can assign values to it by using an index parameter in our constructor.
pd.DataFrame(
{"Apples": [50, 21], "Bananas": [131, 2]}, index=["2018 Sales", "2019 Sales"]
)In essence, it is a single column of a DataFrame, a sequence of data values.
pd.Series([30, 50, 21])You can assign column values to the Series the same way as before, using an index parameter. However, a Series does not have a column name, it only has one overall name.
pd.Series([30, 50, 21], index=["2017 Sales", "2018 Sales", "2019 Sales"], name="Apples")Data can be stored in any of a number of different forms and formats. By far the most basic of these is the humble CSV file. A CSV file is a table of values separated by commas.
# load data
wine_reviews = pd.read_csv(
"../input/wine-reviews/winemag-data-130k-v2.csv", index_col=0
)- If your CSV file has a built-in index, pandas can use that column for the index (instead of creating a new one automatically).
# data dimention
wine_reviews.shape
# columns' name
wine_reviews.columns
# top rows
wine_reviews.head()
# bottom rows
wine_reviews.tail()animals = pd.DataFrame(
{"Cows": [12, 20], "Goats": [22, 19]}, index=["Year 1", "Year 2"]
)
animals.to_csv("cows_and_goats.csv")Pro data scientists do this dozens of times a day. You can, too! #
In Python, we can access the property of an object by accessing it as an attribute. A reviews object, might have a country property, which we can access by calling reviews.country. Columns in a pandas DataFrame work in much the same way.
If we have a Python dictionary, we can access its values using the indexing [] operator.
# select the `country` column
reviews["country"]A pandas Series looks kind of like a dictionary. So, to drill down to a single specific value, we need only use the indexing operator [] once more.
# select the first value from the `country` column
reviews["country"][0]
>>> 'Italy'For more advanced operations, pandas has its own accessor operators, iloc and loc.
Selecting data based on its numerical position in the data, like a matrix.
# select the first row
reviews.iloc[0]
# select the first column, `:` means everything
reviews.iloc[:, 0]
# select the first value from the `country` column
reviews["country"].iloc[0]
# select the last five elements of the dataset
reviews.iloc[-5:]Selecting data based on its index value, with inclusive range.
# select the first value from the `country` column
reviews.loc[0, "country"]
# select all the entries from three specific columns
reviews.loc[:, ["taster_name", "taster_twitter_handle", "points"]]# select first three rows
reviews.iloc[:3]
# or
reviews.loc[:2]# select the first 100 records of the `country` and `variety` columns.
cols_idx = [0, 11]
reviews.iloc[:100, cols_idx]
# or
cols = ["country", "variety"]
reviews.loc[:99, cols]reviews.set_index("title")To do interesting things with the data, we often need to ask questions based on conditions.
To combine multiple conditions in pandas, bitwise operators must be used.
& # AND x & y
| # OR x | y
^ # XOR x ^ y
~ # NOT ~x
>> # right shift x>>
<< # left shift x<<For example, suppose that we're interested in better-than-average wines produced in Italy.
cond1 = reviews["country"] == "Italy"
cond2 = reviews["points"] >= 90
reviews.loc[cond1 & cond2]isin() lets you select data whose value "is in" a list of values.
# select wines only from Italy or France
reviews.loc[reviews["country"].isin(["Italy", "France"])]isna() and notna() let you highlight values which are (or are not) empty (NaN).
# filter out wines lacking a price tag in the dataset
reviews.loc[reviews["price"].notna()]isnull()is an alias forisna(), same asnotnull()fornotna().
# query a df like SQL datasets
wine_reviews.query("price > 100 and country = 'Italy'")# you can assign either a constant value
reviews["critic"] = "everyone"
# or with an iterable of values
reviews["index_backwards"] = range(len(reviews), 0, -1)Extract insights from your data. #
# get summary statistic about a DataFrame
reviews.describe()count: shows how many rows have non-missing values.mean: the average.std: the standard deviation, measures how numerically spread out the values are.min,25%(25th percentile),50%(50th percentiles),75%(75th percentiles) andmax
# get summary statistic about a Series
reviews["points"].describe()
# see the mean
reviews["points"].mean()
# see a list of unique values
reviews["points"].unique()
# see a list of unique values and how often they occur
reviews["points"].value_counts()
# get the titles & points of the 3 highest point
reviews["points"].nlargest(3)# get the title of the wine with the highest points-to-price ratio
max_p2pr = (reviews["points"] / reviews["price"]).idxmax()
reviews.loc[max_p2pr, "title"]
>>> 'Bandit NV Merlot (California)'A function that takes one set of values and "maps" them to another set of values, for creating new representations from existing data.
# remean the scores the wines received to 0
review_points_mean = reviews["points"].mean()
reviews["points"].map(lambda p: p - review_points_mean)# create a series counting how many times each of "tropical" or "fruity" appears in the description column
n_tropical = reviews["description"].map(lambda desc: "tropical" in desc).sum()
n_fruity = reviews["description"].map(lambda desc: "fruity" in desc).sum()
pd.Series([n_tropical, n_fruity], index=["tropical", "fruity"])The function you pass to map() should expect a single value from the Series (a point value, in the above example), and return a transformed version of that value. map() returns a new Series.
# remean the scores the wines received to 0
def remean_points(row):
row["points"] = row["points"] - review_points_mean
return row
reviews.apply(remean_points, axis="columns")# create a series with the number of stars corresponding to each review
def stars(row):
# any wines from country X should automatically get 3 stars, because of ADs' MONEY!
if row["country"] == "X":
return 3
elif row["points"] >= 95:
return 3
elif row["points"] >= 85:
return 2
else:
return 1
reviews.apply(stars, axis="columns")apply() is the equivalent method if we want to transform a whole DataFrame by calling a custom method on each row. apply() returns a new DataFrame.
If we had called reviews.apply() with axis="index", then instead of passing a function to transform each row, we would need to give a function to transform each column.
They perform a simple operation between a lot of values on the left and a single (a lot of) value(s) on the right.
# remean the scores the wines received to 0
review_points_mean = reviews["points"].mean()
reviews["points"] - review_points_mean# combine country and region information in the dataset
reviews["country"] + " - " + reviews["region_1"]These operators are faster than map() or apply() because they uses speed ups built into pandas. All of the standard Python operators (>, <, ==, and so on) work in this manner.
However, they are not as flexible as map() or apply(), which can do more advanced things, like applying conditional logic, which cannot be done with addition and subtraction alone.
Scale up your level of insight. The more complex the dataset, the more this matters. #
Maps allow us to transform data in a DataFrame or Series one value at a time for an entire column. However, often we want to group our data, and then do something specific to the group the data is in.
# replicate what `value_counts()` does
reviews.groupby("points")["points"].count()
# get the minimum price from each group of points
reviews.groupby("points")["price"].min()
# get a series whose index is the taster_twitter_handle values count how many reviews each person wrote
reviews.groupby("taster_twitter_handle").size()
# or
reviews.groupby("taster_twitter_handle")["taster_twitter_handle"].count()
# get the title of the first wine reviewed from each winery
reviews.groupby("winery").apply(lambda df: df["title"].iloc[0])# get a dataframe whose index is the variety category and values are the `min` and `max` prices
reviews.groupby("variety")["price"].agg([min, max])# pick out the best wine by country and province
reviews.groupby(["country", "province"]).apply(lambda df: df.loc[df["points"].idxmax()])Multi-indices have several methods for dealing with their tiered structure which are absent for single-level indices.
They also require two levels of labels to retrieve a value.
The use cases for a multi-index are detailed alongside instructions on using them in the MultiIndex / Advanced Selection section of the pandas documentation.
# convert back to a regular index
count_prov_best.reset_index()# sort (country, province) based on how many reviews are belong to
count_prov_reviewed = reviews.groupby(["country", "province"])["description"].agg([len])
count_prov_reviewed.reset_index().sort_values(by="len", ascending=False)count_prov_reviewed.reset_index().sort_values(by=["country", "len"], ascending=False)# get a series whose index is wine prices and values is the maximum points a wine costing that much was given in a review. sort the values by price, ascending
reviews.groupby("price")["points"].max().sort_index(ascending=True)Deal with the most common progress-blocking problems. #
The data type for a column in a DataFrame or a Series is known as the dtype.
int64,float64,object
# a dataframe
reviews.dtypes
# a series
reviews["price"].dtype
# a dataframe or series index
reviews.index.dtype
# convert a dtype
reviews["points"].astype("float64")# get a series of True & False, based on where NaNs are
reviews["price"].isna()
# find the number of NaNs
reviews["price"].isna().sum()
# create a dataframe of rows with missing country
reviews[reviews["country"].isna()]isnull()is an alias forisna(), same asnotnull()fornotna().
# fill NaNs with Unknown
reviews["region_1"].fillna("Unknown")# replace missing data which is given some kind of sentinel values
reviews["region_1"].replace(["Unknown", "Undisclosed", "Invalid"], "NaN")# filter rows with NaNs
reviews.dropna(axis=0)
# filter columns with NaNs
reviews.dropna(axis=1)Data comes in from many sources. Help it all make sense together. #
# change the names of columns
reviews.rename(columns={"region_1": "region", "region_2": "locale"})
# change the indices of rows
reviews.rename(index={0: "firstEntry", 1: "secondEntry"})# change the names of axes, form rows to wines, from columns to fields
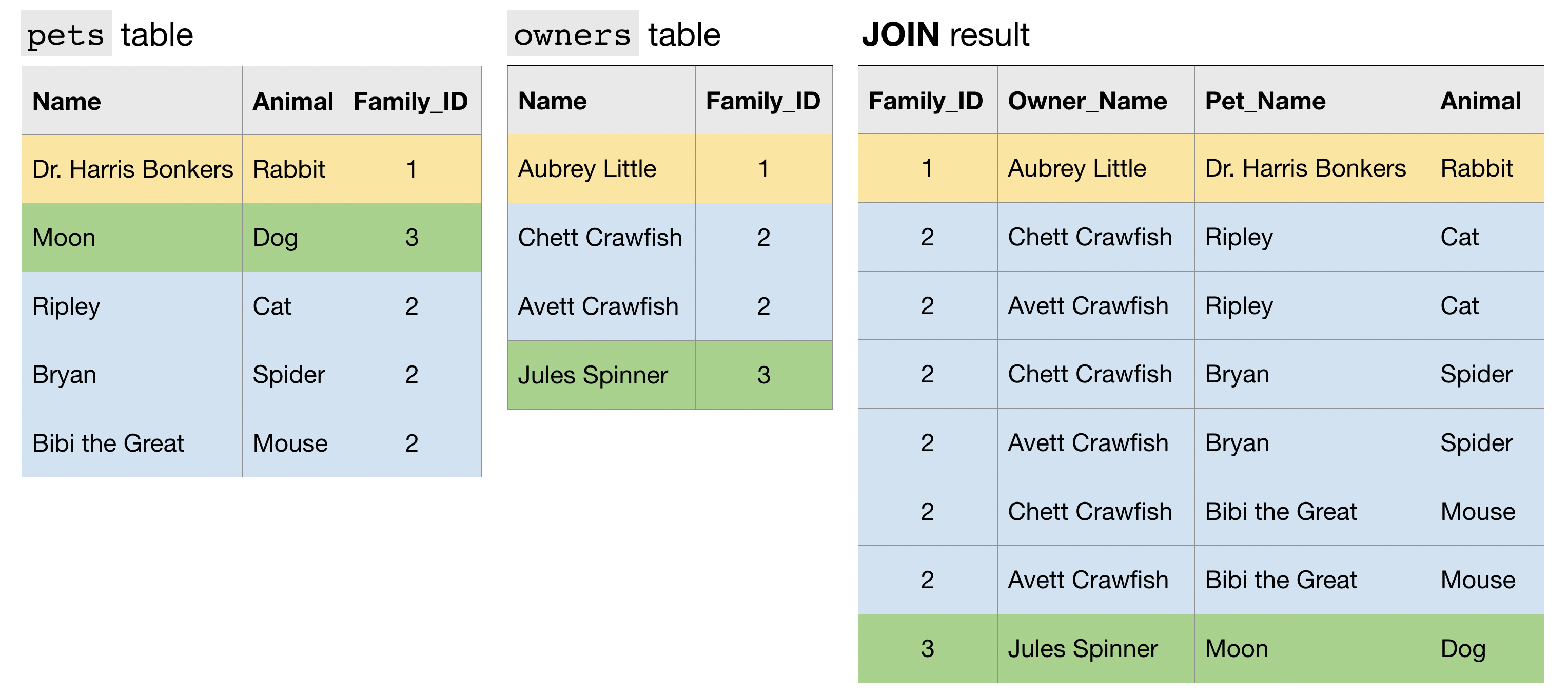
reviews.rename_axis("wines", axis="rows").rename_axis("fields", axis="columns")We will sometimes need to combine different DataFrames and/or Series. Pandas has three core methods for doing this. In order of increasing complexity, these are
It will smush a list of elements together along an axis.
This is useful when we have data in different DataFrame or Series objects but having the same columns.
canadian_yt = pd.read_csv("../input/youtube-new/CAvideos.csv")
british_yt = pd.read_csv("../input/youtube-new/GBvideos.csv")
pd.concat([canadian_yt, british_yt])It lets you combine different DataFrame objects which have an index in common.
# pull down videos that happened to be trending on the same day in both Canada and the UK
left = canadian_yt.set_index(["title", "trending_date"])
right = british_yt.set_index(["title", "trending_date"])
left.join(right, lsuffix="_CAN", rsuffix="_UK")- The
lsuffixandrsuffixparameters are necessary when the data has the same column names in both datasets.
Make great data visualizations. A great way to see the power of coding!
Visualize trends over time. #
import pandas as pd
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns# load a timeseries data file
spotify_data = pd.read_csv("../input/spotify.csv", index_col="Date", parse_dates=True)
# set the width and height of the figure
plt.figure(figsize=(14, 6))
# add title
plt.title("Daily Global Streams of Popular Songs in 2017-2018")
# plot a line chart for daily global streams of each song
sns.lineplot(data=spotify_data)
# plot a subset of the data
sns.lineplot(data=spotify_data["Shape of You"], label="Shape of You")
# add label for horizontal axis
plt.xlabel("Date")Use color or length to compare categories in a dataset. #
# load data
flight_data = pd.read_csv("../input/flight_delays.csv", index_col="Month")
# add title
plt.title("Average Arrival Delay for Spirit Airlines Flights, by Month")
# rotate labels for horizontal axis
plt.xticks(rotation="vertical")
# plot a bar chart, showing average arrival delay for Spirit Airlines flights by month
sns.barplot(x=flight_data.index, y=flight_data["NK"])
# add label for vertical axis
plt.ylabel("Arrival delay (in minutes)")- Note: You must select the indexing column with
flight_data.index, and it is not possible to useflight_data['Month'], because when we loaded the dataset, the"Month"column was used to index the rows.
# add title
plt.title("Average Arrival Delay for Each Airline, by Month")
# plot a heatmap, showing average arrival delay for each airline by month
sns.heatmap(data=flight_data, annot=True)
# add label for horizontal axis
plt.xlabel("Airline")# get the maximum average delay on March
flight_data.loc[3].max()
# find the aireline with the minimum average delay on October
flight_data.loc[10].idxmin()Leverage the coordinate plane to explore relationships between variables. #
# load data
insurance_data = pd.read_csv("../input/insurance.csv")
# a simple scatter plot
sns.scatterplot(x=insurance_data["bmi"], y=insurance_data["charges"])
# add a regression line
sns.regplot(x=insurance_data["bmi"], y=insurance_data["charges"])
# a color-coded scatter plot
sns.scatterplot(
x=insurance_data["bmi"], y=insurance_data["charges"], hue=insurance_data["smoker"]
)
# add two regression lines, corresponding to hue
sns.lmplot(x="bmi", y="charges", hue="smoker", data=insurance_data)
# a categorical scatter plot with non-overlapping points (swarmplot)
sns.swarmplot(x=insurance_data["smoker"], y=insurance_data["charges"])Create histograms and density plots. #
# load data
iris_data = pd.read_csv("../input/iris.csv", index_col="Id")
# a simple histogram
sns.distplot(a=iris_data["Petal Length (cm)"], kde=False)
# a kde (kernel density estimate) plot
sns.kdeplot(data=iris_data["Petal Length (cm)"], shade=True)
# a 2D kde plot
sns.jointplot(
x=iris_data["Petal Length (cm)"], y=iris_data["Sepal Width (cm)"], kind="kde"
)# load data
iris_set_data = pd.read_csv("../input/iris_setosa.csv", index_col="Id")
iris_ver_data = pd.read_csv("../input/iris_versicolor.csv", index_col="Id")
iris_vir_data = pd.read_csv("../input/iris_virginica.csv", index_col="Id")
# kde plots for each one, histograms can be used too
sns.kdeplot(data=iris_set_data["Petal Length (cm)"], label="Setosa", shade=True)
sns.kdeplot(data=iris_ver_data["Petal Length (cm)"], label="Versicolor", shade=True)
sns.kdeplot(data=iris_vir_data["Petal Length (cm)"], label="Virginica", shade=True)
# force legend to appear
plt.legend()Decide how to best tell the story behind your data. #
- A trend is defined as a pattern of change.
sns.lineplot- Line charts are best to show trends over a period of time, and multiple lines can be used to show trends in more than one group.
sns.barplot- Bar charts are useful for comparing quantities corresponding to different groups.sns.heatmap- Heatmaps can be used to find color-coded patterns in tables of numbers.sns.scatterplot- Scatter plots show the relationship between two continuous variables; if color-coded, we can also show the relationship with a third categorical variable.sns.regplot- Including a regression line in the scatter plot makes it easier to see any linear relationship between two variables.sns.lmplot- This command is useful for drawing multiple regression lines, if the scatter plot contains multiple, color-coded groups.sns.swarmplot- Categorical scatter plots show the relationship between a continuous variable and a categorical variable.
- A distribution shows the possible values that we can expect to see in a variable, along with how likely they are.
sns.distplot- Histograms show the distribution of a single numerical variable.sns.kdeplot- KDE plots (or 2D KDE plots) show an estimated, smooth distribution of a single numerical variable (or two numerical variables).sns.jointplot- This command is useful for simultaneously displaying a 2D KDE plot with the corresponding KDE plots for each individual variable.
Practice for real-world application. #
# list all your datasets' folders
import os
print(os.listdir("../input"))How to put your new skills to use for your next personal or work project. #
Create interactive maps, and discover patterns in geospatial data.
Get started with plotting in GeoPandas. #
With this course you can find solutions for several real-world problems like:
- Where should a global non-profit expand its reach in remote areas of the Philippines?
- How do purple martins, a threatened bird species, travel between North and South America? Are the birds travelling to conservation areas?
- Which areas of Japan could potentially benefit from extra earthquake reinforcement?
- Which Starbucks stores in California are strong candidates for the next Starbucks Reserve Roastery location?
- ...
import geopandas as gpdThe data was loaded into a (GeoPandas) GeoDataFrame object has all of the capabilities of a (Pandas) DataFrame. So, every command that you can use with a DataFrame will work with the data!
There are many, many different geospatial file formats, such as shapefile, GeoJSON, KML, and GPKG.
- shapefile is the most common file type that you'll encounter, and
- all of these file types can be quickly loaded with the
read_file()function.
Every GeoDataFrame contains a special geometry column. It contains all of the geometric objects that are displayed when we call the plot() method. While this column can contain a variety of different datatypes, each entry will typically be a Point, LineString, or Polygon.
Create it layer by layer.
# load data
world_loans = gpd.read_file(
"../input/geospatial-learn-course-data/kiva_loans/kiva_loans/kiva_loans.shp"
)
# define a base map with county boundaries
world = gpd.read_file(gpd.datasets.get_path("naturalearth_lowres"))
ax = world.plot(
figsize=(20, 20), color="whitesmoke", linestyle=":", edgecolor="lightgray"
)
# add loans to the base map
world_loans.plot(ax=ax, color="black", markersize=2)You can subset the data for more details.
# subset the data
phl_loans = world_loans.loc[world_loans["country"] == "Philippines"].copy()
# enable fiona driver & load a KML file containing island boundaries
gpd.io.file.fiona.drvsupport.supported_drivers["KML"] = "rw"
phl = gpd.read_file(
"../input/geospatial-learn-course-data/Philippines_AL258.kml", driver="KML"
)
# define a base map with county boundaries
ax_ph = phl.plot(
figsize=(20, 20), color="whitesmoke", linestyle=":", edgecolor="lightgray"
)
# add loans to the base map
phl_loans.plot(ax=ax_ph, color="black", markersize=2)It's pretty amazing that we can represent the Earth's surface in 2 dimensions! #
The world is a three-dimensional globe. So we have to use a map projection method to render it as a flat surface. Map projections can't be 100% accurate. Each projection distorts the surface of the Earth in some way, while retaining some useful property.
- The equal-area projections preserve area.
- The equidistant projections preserve distance.
We use a coordinate reference system (CRS) to show how the projected points correspond to real locations on Earth. CRSs are referenced by European Petroleum Survey Group (EPSG) codes.
When we create a GeoDataFrame from a shapefile, the CRS is already imported for us. But when creating a GeoDataFrame from a CSV file, we have to set the CRS to EPSG 4326, corresponds to coordinates in latitude and longitude.
# create a DataFrame with health facilities in Ghana
import pandas as pd
facilities_df = pd.read_csv(
"../input/geospatial-learn-course-data/ghana/ghana/health_facilities.csv"
)
# convert the DataFrame to a GeoDataFrame
import geopandas as gpd
facilities = gpd.GeoDataFrame(
facilities_df,
geometry=gpd.points_from_xy(facilities_df.Longitude, facilities_df.Latitude),
)
# set the CRS code
facilities.crs = {"init": "epsg:4326"}- We begin by creating a DataFrame containing columns with latitude and longitude coordinates.
- To convert it to a GeoDataFrame, we use
gpd.GeoDataFrame(). - The
gpd.points_from_xy()function creates Point objects from the latitude and longitude columns.
Re-projecting refers to the process of changing the CRS. This is done in GeoPandas with the to_crs() method. For example, when plotting multiple GeoDataFrames, it's important that they all use the same CRS.
# load a GeoDataFrame containing regions in Ghana
regions = gpd.read_file(
"../input/geospatial-learn-course-data/ghana/ghana/Regions/Map_of_Regions_in_Ghana.shp"
)
regions.crs
>>> 32630# create a map
ax = regions.plot(figsize=(8, 8), color="whitesmoke", linestyle=":", edgecolor="black")
facilities.to_crs(epsg=32630).plot(ax=ax, alpha=0.6, markersize=1, zorder=1)In case the EPSG code is not available in GeoPandas, we can change the CRS with what's known as the "proj4 string" of the CRS. The proj4 string to convert to latitude/longitude coordinates is:
# change the CRS to EPSG 4326
regions.to_crs("+proj=longlat +ellps=WGS84 +datum=WGS84 +no_defs")For an arbitrary GeoDataFrame, the type in the geometry column depends on what we are trying to show: for instance, we might use:
- a
Pointfor the epicenter of an earthquake, - a
LineStringfor a street, or - a
Polygonto show country boundaries.
All three types of geometric objects have built-in attributes that you can use to quickly analyze the dataset.
# get the x- or y-coordinates of a point from the x and y attributes
facilities["geometry"].x
# calculate the area (in square kilometers) of all polygons
sum(regions["geometry"].to_crs(epsg=3035).area) / 10 ** 6- ESPG 3035 Scope: Statistical mapping at all scales and other purposes where true area representation is required.
# load data
import pandas as pd
birds_df = pd.read_csv(
"../input/geospatial-learn-course-data/purple_martin.csv", parse_dates=["timestamp"]
)
# create the GeoDataFrame
import geopandas as gpd
birds = gpd.GeoDataFrame(
birds_df,
geometry=gpd.points_from_xy(birds_df["location-long"], birds_df["location-lat"]),
)
# create GeoDataFrame showing path for each bird
from shapely.geometry import LineString
path_df = (
birds.groupby("tag-local-identifier")["geometry"]
.apply(list)
.apply(lambda x: LineString(x))
.reset_index()
)
path_gdf = gpd.GeoDataFrame(path_df, geometry=path_df["geometry"])
path_gdf.crs = {"init": "epsg:4326"}Learn how to make interactive heatmaps, choropleth maps, and more! #
# load data
import pandas as pd
crimes = pd.read_csv(
"../input/geospatial-learn-course-data/crimes-in-boston/crimes-in-boston/crime.csv",
encoding="latin-1",
)
# drop rows with missing locations
crimes.dropna(subset=["Lat", "Long", "DISTRICT"], inplace=True)
# focus on major crimes in 2018
crimes = crimes[
crimes["OFFENSE_CODE_GROUP"].isin(
[
"Larceny",
"Auto Theft",
"Robbery",
"Larceny From Motor Vehicle",
"Residential Burglary",
"Simple Assault",
"Harassment",
"Ballistics",
"Aggravated Assault",
"Other Burglary",
"Arson",
"Commercial Burglary",
"HOME INVASION",
"Homicide",
"Criminal Harassment",
"Manslaughter",
]
)
]
crimes = crimes[crimes["YEAR"] == 2018]
# focus on daytime robberies
daytime_robberies = crimes[
((crimes["OFFENSE_CODE_GROUP"] == "Robbery") & (crimes["HOUR"].isin(range(9, 18))))
]In this tutorial, you'll learn how to create interactive maps with the folium package. We create the base map with folium.Map().
# create the base map
from folium import Map
base_map = Map(location=[42.32, -71.0589], tiles="openstreetmap", zoom_start=10)locationsets the initial center of the map. We use the latitude (42.32° N) and longitude (-71.0589° E) of the city of Boston.tileschanges the styling of the map; in this case, we choose the OpenStreetMap style. If you're curious, you can find the other options listed here.zoom_startsets the initial level of zoom of the map, where higher values zoom in closer to the map.
We add markers to the map with folium.Marker(). Each marker below corresponds to a different robbery.
# define the base map
map_marker = map_base
# add points to the map
from folium import Marker
for idx, row in daytime_robberies.iterrows():
Marker([row["Lat"], row["Long"]], popup=row["HOUR"]).add_to(map_marker)
# display the map
map_markerIf we have a lot of markers to add, folium.plugins.MarkerCluster() can help to declutter the map. Each marker is added to a MarkerCluster object.
# define the base map
map_cluser = map_base
# add points to the map
from folium.plugins import MarkerCluster
mc = MarkerCluster()
from folium import Marker
import math
for idx, row in daytime_robberies.iterrows():
if not math.isnan(row["Lat"]) and not math.isnan(row["Long"]):
mc.add_child(Marker([row["Lat"], row["Long"]]))
map_cluser.add_child(mc)
# display the map
map_cluser- We used
math.isnan()becauserow["Lat"]orrow["Long"]arefloat.
A bubble map uses circles instead of markers. By varying the size and color of each circle, we can also show the relationship between location and two other variables.
We create a bubble map by using folium.Circle() to iteratively add circles.
# define the base map
map_bubble = map_base
# define color/size producer function
def color_producer(val):
if val <= 12:
# robberies that occurred in hours 9-12
return "forestgreen"
else:
# robberies from hours 13-17
return "darkred"
# add a bubble map to the base map
from folium import Circle
for i in range(len(daytime_robberies)):
Circle(
location=[daytime_robberies.iloc[i]["Lat"], daytime_robberies.iloc[i]["Long"]],
radius=20,
color=color_producer(daytime_robberies.iloc[i]["HOUR"]),
).add_to(map_bubble)
# display the map
map_bubblelocationis a list containing the center of the circle, in latitude and longitude.radiussets the radius of the circle.- We can implement this by defining a function similar to the
color_producer()function that is used to vary the color of each circle.
- We can implement this by defining a function similar to the
colorsets the color of each circle.The color_producer()function is used to visualize the effect of the hour on robbery location.
To create a heatmap, we use folium.plugins.HeatMap(). This shows the density of crime in different areas of the city, where red areas have relatively more criminal incidents.
# define the base map
map_heat = map_base
# add a heatmap to the base map
from folium.plugins import HeatMap
HeatMap(data=crimes[["Lat", "Long"]], radius=10).add_to(map_heat)
# display the map
map_heatdatais a DataFrame containing the locations that we'd like to plot.radiuscontrols the smoothness of the heatmap. Higher values make the heatmap look smoother.
To understand how crime varies by police district, we'll create a choropleth map. To create a choropleth, we use folium.Choropleth().
As a first step, we create a GeoDataFrame where each district is assigned a different row, and the geometry column contains the geographical boundaries.
# create GeoDataFrame with geographical boundaries of districts
import geopandas as gpd
districts_full = gpd.read_file(
"../input/geospatial-learn-course-data/Police_Districts/Police_Districts/Police_Districts.shp"
)
districts = districts_full[["DISTRICT", "geometry"]].set_index("DISTRICT")# create a Pandas Series shows the number of crimes in each police district
plot_dict = crimes["DISTRICT"].value_counts()- It's very important that
plot_dicthas the same index as districts - this is how the code knows how to match the geographical boundaries with appropriate colors.
# define the base map
map_choropleth = map_base
# add a choropleth map to the base map
from folium import Choropleth
Choropleth(
geo_data=districts.__geo_interface__,
data=plot_dict,
key_on="feature.id",
fill_color="YlGnBu",
legend_name="Major Criminal Incidents (Jan-Aug 2018)",
).add_to(map_choropleth)
# display the map
map_choroplethgeo_datais a GeoJSON FeatureCollection containing the boundaries of each geographical area.- We convert the districts GeoDataFrame to a GeoJSON FeatureCollection with the
__geo_interface__attribute.
- We convert the districts GeoDataFrame to a GeoJSON FeatureCollection with the
datais a Pandas Series containing the values that will be used to color-code each geographical area.key_onwill always be set tofeature.id, based on the GeoJSON structure.fill_colorsets the color scale.
Find locations with just the name of a place. And, learn how to join data based on spatial relationships. #
Geocoding is the process of converting the name of a place or an address to a location on a map. We'll use geopandas.tools.geocode() to do all of our geocoding.
from geopandas.tools import geocode
geocode("The Great Pyramid of Giza", provider="nominatim")To use the geocoder, we need:
- the
nameoraddressas a Python string, and - the name of the
provider. To avoid having to provide an API key, we used the OpenStreetMap Nominatim geocoder.
It's often the case that we'll need to geocode many different addresses.
# load Starbucks locations in California
import pandas as pd
starbucks = pd.read_csv("../input/geospatial-learn-course-data/starbucks_locations.csv")# define geocoder function
def my_geocoder(row):
try:
point = geocode(row, provider="nominatim")["geometry"][0]
return pd.Series({"Latitude": point.y, "Longitude": point.x})
except:
return NoneIf the geocoding is successful, it returns a GeoDataFrame with two columns:
- the
geometrycolumn, which is aPointobject, and we can get theLatitudeandLongitudefrom theyandxattributes, respectively. - the
addresscolumn contains the full address.
When geocoding a large dataframe, you might encounter an error when geocoding.
- In case you get a time out error, try first using the
timeoutparameter (allow the service a bit more time to respond). - In case of Too Many Requests error, you have hit the rate-limit of the service, and you should slow down your requests.
GeoPyprovides additional tools for taking into account rate limits in geocoding services. More info.
# rows with missing locations
rows_with_missing = starbucks[
starbucks["Latitude"].isna() | starbucks["Longitude"].isna()
]# fill missing geo data
rows_with_missing = rows_with_missing.apply(lambda x: my_geocoder(x["Address"]), axis=1)
# drop rows that were not successfully geocoded
rows_with_missing.dropna(axis=0, subset=["Latitude", "Longitude"])
# update main DataFrame
starbucks.update(rows_with_missing)We can combine data from different sources.
You already know how to use pd.DataFrame.join() to combine information from multiple DataFrames with a shared index. We refer to this way of joining data (by simpling matching values in the index) as an attribute join. We'll work with some DataFrames containing data and a unique id (in the GEOID column) for each county in the state of California.
# create DataFrame contains an estimate of the population of each county
CA_pop = pd.read_csv(
"../input/geospatial-learn-course-data/CA_county_population.csv", index_col="GEOID"
)
# create DataFrame contains the number of households with high income
CA_high_earners = pd.read_csv(
"../input/geospatial-learn-course-data/CA_county_high_earners.csv",
index_col="GEOID",
)
# create DataFrame contains the median age for each county
CA_median_age = pd.read_csv(
"../input/geospatial-learn-course-data/CA_county_median_age.csv", index_col="GEOID"
)# use an attribute join
cols_to_add = CA_pop.join([CA_high_earners, CA_median_age]).reset_index()When performing an attribute join with a GeoDataFrame, it's best to use the gpd.GeoDataFrame.merge(). We'll work with a GeoDataFrame CA_counties containing the name, area (in square kilometers), and a unique id (in the GEOID column) for each county in the state of California. The geometry column contains a polygon with county boundaries.
import geopandas as gpd
CA_counties = gpd.read_file(
"../input/geospatial-learn-course-data/CA_county_boundaries/CA_county_boundaries/CA_county_boundaries.shp"
)# use an attribute join
CA_stats = CA_counties.merge(cols_to_add, on="GEOID")- The
onargument is set to the column name that is used to match rows.
Now that we have all of the data in one place, it's much easier to calculate statistics that use a combination of columns.
CA_stats["density"] = CA_stats["population"] / CA_stats["area_sqkm"]With a spatial join, we combine GeoDataFrames based on the spatial relationship between the objects in the geometry columns. We do this with gpd.sjoin().
So, which counties look promising for new Starbucks Reserve Roastery?
sel_counties = CA_stats[
(CA_stats["high_earners"] >= 100000)
& (CA_stats["median_age"] <= 38.5)
& (CA_stats["density"] >= 285)
]
sel_counties.crs = {"init": "epsg:4326"}starbucks_gdf = gpd.GeoDataFrame(
starbucks,
geometry=gpd.points_from_xy(starbucks["Longitude"], starbucks["Latitude"]),
)
starbucks_gdf.crs = {"init": "epsg:4326"}sel_counties_stores = gpd.sjoin(starbucks_gdf, sel_counties)The spatial join above looks at the geometry columns in both GeoDataFrames. If a Point object from the starbucks_gdf GeoDataFrame intersects a Polygon object from the sel_counties DataFrame, the corresponding rows are combined and added as a single row of the sel_counties_stores DataFrame. Otherwise, counties without a matching starbuckses (and starbuckses without a matching county) are omitted from the results.
The gpd.sjoin() method is customizable for different types of joins, through the how and op arguments. For example, you can do the equivalent of a SQL left (or right) join by setting how='left' (or how='right').
Let's visualize!
# define the base map
from folium import Map
map_cluser = Map(location=[37, -120], zoom_start=6)
# add points to the map
from folium.plugins import MarkerCluster
mc = MarkerCluster()
from folium import Marker
for idx, row in sel_counties_stores.iterrows():
mc.add_child(Marker([row["Latitude"], row["Longitude"]]))
map_cluser.add_child(mc)
# display the map
map_cluserMeasure distance, and explore neighboring points on a map. #
We'll explore several techniques for proximity analysis, such as:
- Measuring the distance between points on a map, and
- Selecting all points within some radius of a feature.
We want to identify how hospitals have been responding to crash collisions in New York City. We'll work with GeoDataFrame collisions tracking major motor vehicle collisions in 2013-2018.
import geopandas as gpd
collisions = gpd.read_file(
"../input/geospatial-learn-course-data/NYPD_Motor_Vehicle_Collisions/NYPD_Motor_Vehicle_Collisions/NYPD_Motor_Vehicle_Collisions.shp"
)
hospitals = gpd.read_file(
"../input/geospatial-learn-course-data/nyu_2451_34494/nyu_2451_34494/nyu_2451_34494.shp"
)To measure distances between points from two different GeoDataFrames, we first have to make sure that they use the same CRS.
collisions.crs == hospitals.crs- We also check the CRS to see which units it uses (meters, feet, or something else). In this case, EPSG 2263 has units of meters.
Then, we use the distance() method, returns a Series containing the distance to the others.
# measure distance from a relatively recent collision to each hospital
distances = hospitals["geometry"].distance(collisions.iloc[-1]["geometry"])# calculate mean distance to hospitals
distances.mean()
# find the closest hospital
hospitals.iloc[distances.idxmin()][["ADDRESS", "LATITUDE", "LONGITUDE"]]If we want to understand all points on a map that are some radius away from a point, the simplest way is to create a buffer. It's a GeoSeries containing multiple Polygon objects. Each polygon is a buffer around a different spot.
We'll create a DataFrame outside_range containing all rows from collisions with crashes that occurred more than 10 kilometers from the closest hospital.
# create a GeoSeries buffer
ten_km_buffer = hospitals["geometry"].buffer(10000)To test if a collision occurred within 10 kilometers of any hospital, we could run different tests for each polygon. But a more efficient way is to first collapse all of the polygons into a MultiPolygon object. We do this with the unary_union attribute.
# turn group of polygons into single multipolygon
my_union = ten_km_buffer.unary_unionWe use the contains() method to check if the multipolygon contains a point.
# is the closest station less than two miles away?
my_union.contains(collisions.iloc[-1]["geometry"])outside_range = collisions.loc[
~collisions["geometry"].apply(lambda x: my_union.contains(x))
]# calculate the percentage of collisions occurred more than 10 km away from the closest hospital
round(100 * len(outside_range) / len(collisions), 2)When collisions occur in distant locations, it becomes even more vital that injured persons are transported to the nearest available hospital.
With this in mind, we want to create a recommender that:
- takes the location of the crash as input,
- finds the closest hospital, and
- returns the name of the closest hospital.
def best_hospital(collision_location):
idx_min = hospitals["geometry"].distance(collision_location).idxmin()
return hospitals.iloc[idx_min]["name"]# suggest the closest hospital to the last collision
best_hospital(outside_range["geometry"].iloc[-1])# which hospital is most recommended?
outside_range["geometry"].apply(best_hospital).value_counts().idxmax()Where should the city construct new hospitals? Lets visualize!
# define the base map
from folium import Map
m = Map(location=[40.7, -74], zoom_start=11)
# add buffers' Polygon
from folium import GeoJson
GeoJson(ten_km_buffer.to_crs(epsg=4326)).add_to(m)
# add the heatmap of collisions, out of 10km buffers
from folium.plugins import HeatMap
HeatMap(data=outside_range[["LATITUDE", "LONGITUDE"]], radius=9).add_to(m)
# add (Lat,Long) popup
from folium import LatLngPopup
LatLngPopup().add_to(m)
# display the map
m- We use
folium.GeoJson()to plot eachPolygonon a map. Note that since folium requires coordinates in latitude and longitude, we have to convert the CRS to EPSG 4326 before plotting.
Learn the core ideas in machine learning, and build your first models.
The first step if you're new to machine learning. #
- Fitting or Training: Capturing patterns from training data
- Predicting: Getting results from applying the model to new data
Load and understand your data. #
# load data
import pandas as pd
melbourne_data = pd.read_csv("../input/melbourne-housing-snapshot/melb_data.csv")# summary
melbourne_data.head()
melbourne_data.describe()Building your first model. Hurray! #
# filter rows with missing values
dropna_melbourne_data = melbourne_data.dropna(axis=0)# separate target (y) from features (predictors, X)
y = dropna_melbourne_data["Price"]
feature_list = [
"Rooms",
"Bathroom",
"Landsize",
"BuildingArea",
"YearBuilt",
"Lattitude",
"Longtitude",
]
X = dropna_melbourne_data[feature_list]- Define: What type of model will it be? A decision tree? Some other type of model?
- Fit: Capture patterns from provided data. This is the heart of modeling.
- Predict: Just what it sounds like.
- Evaluate: Determine how accurate the model's predictions are.
# define model
from sklearn.tree import DecisionTreeRegressor
melbourne_model = DecisionTreeRegressor(random_state=1)
# fit model
melbourne_model.fit(X, y)
# make prediction
predictions = melbourne_model.predict(X)Measure the performance of your model? So you can test and compare alternatives. #
There are many metrics for summarizing the model quality. Predictive accuracy means will the model's predictions be close to what actually happens?
Mean Absolute Error (MAE)
from sklearn.metrics import mean_absolute_error
mean_absolute_error(y, predictions)Big Mistake: Measuring scores with the training data or the problem with in-sample scores!
Making predictions on new data
Exclude some data from the model-building process, and then use those to test the model's accuracy.
# break off validation set from training data, for both features and target
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(X, y, random_state=1)# define model
melbourne_model = DecisionTreeRegressor(random_state=1)
# fit model
melbourne_model.fit(X_train, y_train)
# make prediction on validation data
predictions_val = melbourne_model.predict(X_valid)
# evaluate the model
mean_absolute_error(y_valid, predictions_val)There are many ways to improve a model, such as
- Finding better features, the iterating process of building models with different features and comparing them to each other
- Finding better model types
Fine-tune your model for better performance. #
- Overfitting: Capturing spurious patterns that won't recur in the future, leading to less accurate predictions.
- Underfitting: Failing to capture relevant patterns, again leading to less accurate predictions.
In the Decision Tree model, the most important option to control the accuracy is the tree's depth, a measure of how many splits it makes before coming to a prediction.
- A deep tree makes leaves with fewer objects. It causes overfitting.
- A shallow tree makes big groups. It causes underfitting.
There are a few options for controlling the tree depth, and many allow for some routes through the tree to have greater depth than other routes. But the max_leaf_nodes argument provides a very sensible way to control overfitting vs underfitting.
# function for comparing MAE with differing values of max_leaf_nodes
def get_mae(max_leaf_nodes, X_train, X_valid, y_train, y_valid):
model = DecisionTreeRegressor(max_leaf_nodes=max_leaf_nodes, random_state=0)
model.fit(X_train, y_train)
predictions_val = model.predict(X_valid)
mae = mean_absolute_error(y_valid, predictions_val)
return mae# compare models
max_leaf_nodes_candidates = [5, 50, 500, 5000]
scores = {
leaf_size: get_mae(leaf_size, X_train, X_valid, y_train, y_valid)
for leaf_size in max_leaf_nodes_candidates
}
best_tree_size = min(scores, key=scores.get)Using a more sophisticated machine learning algorithm. #
Decision trees leave you with a difficult decision. A deep tree and overfitting vs. a shallow one and underfitting.
A Random Forest model uses many trees, and makes a prediction by averaging the predictions of each component. It generally has much better predictive accuracy even with than a single decision tree, even with default parameters, without tuning the parameters like max_leaf_nodes.
# define & fit model
from sklearn.ensemble import RandomForestRegressor
forest_model = RandomForestRegressor(random_state=1)
forest_model.fit(X_train, y_train)
# make prediction
preds_valid = forest_model.predict(X_valid)
# evaluate the model
from sklearn.metrics import mean_absolute_error
mean_absolute_error(y_valid, preds_valid)Some models, like the XGBoost model, provides better performance when tuned well with the right parameters (but which requires some skill to get the right model parameters).
Enter the world of machine learning competitions to keep improving and see your progress. #
# load data
import pandas as pd
X_full = pd.read_csv("../input/train.csv", index_col="Id")
X_test_full = pd.read_csv("../input/test.csv", index_col="Id")# separate target (y) from features (X)
y = X_full["SalePrice"]
features = [
"LotArea",
"YearBuilt",
"1stFlrSF",
"2ndFlrSF",
"FullBath",
"BedroomAbvGr",
"TotRmsAbvGrd",
]
X = X_full[features].copy()
X_test = X_test_full[features].copy()# break off validation set from training data
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=0
)# define models
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
model_1 = DecisionTreeRegressor(random_state=0)
model_2 = DecisionTreeRegressor(max_leaf_nodes=100, random_state=0)
model_3 = RandomForestRegressor(n_estimators=50, random_state=0)
model_4 = RandomForestRegressor(n_estimators=100, random_state=0)
model_5 = RandomForestRegressor(n_estimators=100, criterion="mae", random_state=0)
model_6 = RandomForestRegressor(n_estimators=200, min_samples_split=20, random_state=0)
model_7 = RandomForestRegressor(n_estimators=100, max_depth=7, random_state=0)
models = [model_1, model_2, model_3, model_4, model_5, model_6, model_7]# function for comparing different models
from sklearn.metrics import mean_absolute_error
def score_model(model, X_train, X_valid, y_train, y_valid):
# fit model
model.fit(X_train, y_train)
# make validation predictions
preds_valid = model.predict(X_valid)
# return mae
return mean_absolute_error(y_valid, preds_valid)# compare models
for i in range(len(models)):
mae = score_model(models[i])
print(f"Model {i+1} MAE: {mae:,.0f}")Model 1 MAE: 29,653
Model 2 MAE: 27,283
Model 3 MAE: 24,015
Model 4 MAE: 23,740
Model 5 MAE: 23,528
Model 6 MAE: 23,996
Model 7 MAE: 23,706# define model, based on the most accurate model
my_model = RandomForestRegressor(n_estimators=100, criterion="mae", random_state=0)
# fit the model to the training data, all of it
my_model.fit(X, y)
# make test prediction
preds_test = my_model.predict(X_test)# save predictions in format used for competition scoring
output = pd.DataFrame({"Id": X_test.index, "SalePrice": preds_test})
output.to_csv("submission.csv", index=False)Learn to handle missing values, non-numeric values, data leakage and more. Your models will be more accurate and useful.
Review what you need for this Micro-Course. #
In this micro-course, you will accelerate your machine learning expertise by learning how to:
- Tackle data types often found in real-world datasets (missing values, categorical variables),
- Design pipelines to improve the quality of your machine learning code,
- Use advanced techniques for model validation (cross-validation),
- Build state-of-the-art models that are widely used to win Kaggle competitions (XGBoost), and
- Avoid common and important data science mistakes (leakage).
Missing values happen. Be prepared for this common challenge in real datasets. #
There are many ways data can end up with missing values. For example,
- A 2 bedroom house won't include a value for the size of a third bedroom.
- A survey respondent may choose not to share his income.
Most machine learning libraries (including scikit-learn) give an error if you try to build a model using data with missing values.
# show number of missing values in each column
def missing_val_count(data):
missing_val_count_by_column = data.isna().sum()
return missing_val_count_by_column[missing_val_count_by_column > 0]- Drop Columns with Missing Values
- Imputation
# load data
import pandas as pd
X_full = pd.read_csv("../input/train.csv", index_col="Id")
X_test_full = pd.read_csv("../input/test.csv", index_col="Id")# remove rows with missing "SalePrice"
X_full.dropna(axis=0, subset=["SalePrice"], inplace=True)# separate target (y) from features (X)
y = X_full["SalePrice"]
X_full.drop(["SalePrice"], axis=1, inplace=True)# use only numerical features, to keep things simple
X = X_full.select_dtypes(exclude=["object"])
X_test = X_test_full.select_dtypes(exclude=["object"])# break off validation set from training data
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=0
)# get names of columns with missing values
cols_with_missing = [col for col in X_train.columns if X_train[col].isna().any()]# function for comparing different approaches
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
def score_dataset(X_train, X_valid, y_train, y_valid):
model = RandomForestRegressor(n_estimators=10, random_state=0)
model.fit(X_train, y_train)
preds_valid = model.predict(X_valid)
return mean_absolute_error(y_valid, preds_valid)The model loses access to a lot of (potentially useful!) information with this approach.
# drop `cols_with_missing` in training and validation data
reduced_X_train = X_train.drop(cols_with_missing, axis=1)
reduced_X_valid = X_valid.drop(cols_with_missing, axis=1)
# evaluate the model
score_dataset(reduced_X_train, reduced_X_valid, y_train, y_valid)Imputation fills in the missing values with some number.
# imputation
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(strategy="mean")
imputed_X_train = pd.DataFrame(imputer.fit_transform(X_train))
imputed_X_valid = pd.DataFrame(imputer.transform(X_valid))
# imputation removed column names; put them back
imputed_X_train.columns = X_train.columns
imputed_X_valid.columns = X_valid.columns
# evaluate the model
score_dataset(imputed_X_train, imputed_X_valid, y_train, y_valid)Strategy
- default=
meanreplaces missing values using the mean along each column. (only numeric) medianreplaces missing values using the median along each column. (only numeric)most_frequentreplaces missing using the most frequent value along each column. (strings or numeric)constantreplaces missing values withfill_value. (strings or numeric)
# define and fit model
model = RandomForestRegressor(n_estimators=100, random_state=0)
model.fit(imputed_X_train, y_train)
# make validation prediction
preds_valid = model.predict(imputed_X_valid)
mean_absolute_error(y_valid, preds_valid)# preprocess test data
imputed_X_test = pd.DataFrame(imputer.fit_transform(X_test))
# put column names back
imputed_X_test.columns = X_test.columns
# make test prediction
preds_test = model.predict(imputed_X_test)# save test predictions to file
output = pd.DataFrame({"Id": X_test.index, "SalePrice": preds_test})
output.to_csv("submission.csv", index=False)There's a lot of non-numeric data out there. Here's how to use it for machine learning. #
A categorical variable takes only a limited number of values.
- Ordinal: A question that asks "how often you eat breakfast?" and provides four options: "Never", "Rarely", "Most days", or "Every day".
- Nominal: A question that asks "what brand of car you own?".
Most machine learning libraries (including scikit-learn) give an error if you try to build a model using data with categorical variables.
- Drop Categorical Variables
- Label Encoding
- One-Hot Encoding
# load data
import pandas as pd
X_full = pd.read_csv("../input/train.csv", index_col="Id")
X_test_full = pd.read_csv("../input/test.csv", index_col="Id")# remove rows with missing target
X_full.dropna(axis=0, subset=["SalePrice"], inplace=True)# separate target (y) from features (X)
y = data["Price"]
X = data.drop(["Price"], axis=1)# break off validation set from training data
from sklearn.model_selection import train_test_split
X_train_full, X_valid_full, y_train, y_valid = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=0
)# handle missing values (simplest approach)
cols_with_missing = [
col for col in X_train_full.columns if X_train_full[col].isna().any()
]
X_train_full.drop(cols_with_missing, axis=1, inplace=True)
X_valid_full.drop(cols_with_missing, axis=1, inplace=True)# select categorical columns with relatively low cardinality, to keep things simple
# cardinality means the number of unique values in a column
categorical_cols = [
cname
for cname in X_train_full.columns
if (X_train_full[cname].dtype == "object") and (X_train_full[cname].nunique() < 10)
]
# select numerical columns
numerical_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].dtype in ["int64", "float64"]
]
# keep selected columns only
my_cols = categorical_cols + numerical_cols
X_train = X_train_full[my_cols].copy()
X_valid = X_valid_full[my_cols].copy()
X_test = X_test_full[my_cols].copy()# function for comparing different approaches
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
def score_dataset(X_train, X_valid, y_train, y_valid):
model = RandomForestRegressor(n_estimators=100, random_state=0)
model.fit(X_train, y_train)
preds = model.predict(X_valid)
return mean_absolute_error(y_valid, preds)This approach will only work well if the columns did not contain useful information.
# drop catagorial columns
drop_X_train = X_train.select_dtypes(exclude=["object"])
drop_X_valid = X_valid.select_dtypes(exclude=["object"])
# evaluate the model
score_dataset(drop_X_train, drop_X_valid, y_train, y_valid)Label encoding assigns each unique value, that appears in the training data, to a different integer.
In the case that the validation data contains values that don't also appear in the training data, the encoder will throw an error, because these values won't have an integer assigned to them. It should be used only for target labels encoding.
To encode categorical features, use One-Hot Encoder, which can handle unseen values.
For tree-based models (like decision trees and random forests), you can expect label encoding to work well with ordinal variables.
# find columns, which are in validation data but not in training data
good_label_cols = [
col for col in categorical_cols if set(X_train[col]) == set(X_valid[col])
]
bad_label_cols = list(set(categorical_cols) - set(good_label_cols))
# drop them
label_X_train = X_train.drop(bad_label_cols, axis=1)
label_X_valid = X_valid.drop(bad_label_cols, axis=1)
# apply label encoder
from sklearn.preprocessing import LabelEncoder
label_encoder = LabelEncoder()
for col in good_label_cols:
label_X_train[col] = label_encoder.fit_transform(X_train[col])
label_X_valid[col] = label_encoder.transform(X_valid[col])
# evaluate the model
score_dataset(label_X_train, label_X_valid, y_train, y_valid)One-hot encoding creates new columns indicating the presence (or absence) of each possible value in the original data. Useful parameters are:
handle_unknown="ignore"avoids errors when the validation data contains classes that aren't represented in the training data,sparse=Falsereturns the encoded columns as a numpy array (instead of a sparse matrix).
In contrast to label encoding, one-hot encoding does not assume an ordering of the categories. Thus, you can expect this approach to work particularly well with categorical variables without an intrinsic ranking, we refer them as nominal variables.
One-hot encoding generally does not perform well with high-cardinality categorical variable (i.e., more than 15 different values). Cardinality means the number of unique values in a column.
# get cardinality for each column with categorical data
object_nunique = list(map(lambda col: X_train[col].nunique(), categorical_cols))
d = dict(zip(categorical_cols, object_nunique))
# print cardinality by column, in ascending order
sorted(d.items(), key=lambda x: x[1])For this reason, we typically will only one-hot encode columns with relatively low cardinality. Then, high cardinality columns can either be dropped from the dataset, or we can use label encoding.
# columns that will be one-hot encoded
low_cardinality_cols = [col for col in categorical_cols if X_train[col].nunique() < 10]
# columns that will be dropped from the dataset
high_cardinality_cols = list(set(categorical_cols) - set(low_cardinality_cols))
# apply one-hot encoder to each column with categorical data
from sklearn.preprocessing import OneHotEncoder
oh_encoder = OneHotEncoder(handle_unknown="ignore", sparse=False)
oh_cols_train = pd.DataFrame(oh_encoder.fit_transform(X_train[low_cardinality_cols]))
oh_cols_valid = pd.DataFrame(oh_encoder.transform(X_valid[low_cardinality_cols]))
# one-hot encoding removed index; put it back
oh_cols_train.index = X_train.index
oh_cols_valid.index = X_valid.index
# drop all categorical columns (will replace with one-hot encoding)
num_X_train = X_train.drop(categorical_cols, axis=1)
num_X_valid = X_valid.drop(categorical_cols, axis=1)
# add one-hot encoded columns to numerical features
oh_X_train = pd.concat([num_X_train, oh_cols_train], axis=1)
oh_X_valid = pd.concat([num_X_valid, oh_cols_valid], axis=1)
# evaluate the model
score_dataset(oh_X_train, oh_X_valid, y_train, y_valid)Doing all things seperately for training, evaluating and testing is way DIFFICULT. Doing with Pipelines is FUN!
A critical skill for deploying (and even testing) complex models with pre-processing. #
Pipelines are a simple way to keep your data preprocessing and modeling code organized. Specifically, a pipeline bundles preprocessing and modeling steps so you can use the whole bundle as if it were a single step.
Some important benefits of pipelines are:
- Cleaner Code: Accounting for data at each step of preprocessing can get messy. With a pipeline, you won't need to manually keep track of your training and validation data at each step.
- Fewer Bugs: There are fewer opportunities to misapply a step or forget a preprocessing step.
- Easier to Productionize: It can be surprisingly hard to transition a model from a prototype to something deployable at scale, but pipelines can help.
- More Options for Model Validation: You will see an example in the Cross-Validation tutorial.
- Setup
- Define Preprocessing Steps
- Define the Model
- Create and Evaluate the Pipeline
# load data
import pandas as pd
X_full = pd.read_csv("../input/train.csv", index_col="Id")
X_test_full = pd.read_csv("../input/test.csv", index_col="Id")# remove rows with missing target
X_full.dropna(axis=0, subset=["SalePrice"], inplace=True)# separate target (y) from features (X)
y = X_full["Price"]
X = X_full.drop(["Price"], axis=1)# break off validation set from training data
from sklearn.model_selection import train_test_split
X_train_full, X_valid_full, y_train, y_valid = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=0
)# select categorical columns with relatively low cardinality, to keep things simple
# cardinality means the number of unique values in a column
categorical_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].nunique() < 10 and X_train_full[cname].dtype == "object"
]
# select numerical columns
numerical_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].dtype in ["int64", "float64"]
]
# keep selected columns only
my_cols = categorical_cols + numerical_cols
X_train = X_train_full[my_cols].copy()
X_valid = X_valid_full[my_cols].copy()
X_test = X_test_full[my_cols].copy()Similar to how a pipeline bundles together preprocessing and modeling steps, we use the ColumnTransformer class to bundle together different preprocessing steps. The code below:
- Imputes missing values in numerical data, and
- Imputes missing values and applies a one-hot encoding to categorical data.
from sklearn.pipeline import Pipeline
# preprocessing for numerical data
from sklearn.impute import SimpleImputer
numerical_transformer = SimpleImputer(strategy="most_frequent")
# preprocessing for categorical data
from sklearn.preprocessing import OneHotEncoder
categorical_transformer = Pipeline(
steps=[
("imputer", SimpleImputer(strategy="most_frequent")),
("onehot", OneHotEncoder(handle_unknown="ignore")),
]
)
# bundle preprocessing for numerical and categorical data
from sklearn.compose import ColumnTransformer
preprocessor = ColumnTransformer(
transformers=[
("num", numerical_transformer, numerical_cols),
("cat", categorical_transformer, categorical_cols),
]
)For example we use RandomForestRegressor
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor(n_estimators=100, min_samples_split=3, random_state=0)We use the Pipeline class to define a pipeline that bundles the preprocessing and modeling steps.
- With the pipeline, we preprocess the training data and fit the model in a single line of code.
- With the pipeline, we supply the unprocessed features in X_valid to the predict() command, and the pipeline automatically preprocesses the features before generating predictions.
# bundle preprocessing and modeling code in a pipeline
my_pipeline = Pipeline(steps=[("preprocessor", preprocessor), ("model", model)])
# preprocessing of training data, fit model
my_pipeline.fit(X_train, y_train)
# preprocessing of validation data, get predictions
preds = my_pipeline.predict(X_valid)
# evaluate the model
from sklearn.metrics import mean_absolute_error
mean_absolute_error(y_valid, preds)# preprocessing of test data, fit model
preds_test = my_pipeline.predict(X_test)
# save predictions in format used for competition scoring
output = pd.DataFrame({"Id": X_test.index, "SalePrice": preds_test})
output.to_csv("submission.csv", index=False)Vola!
A better way to test your models. #
Machine learning is an iterative process. You will face choices about what predictive variables to use, what types of models to use, what arguments to supply to those models, etc.
In a dataset with 5000 rows, you will typically keep about 20% of the data as a validation dataset, or 1000 rows. But this leaves some random chance in determining model scores. That is, a model might do well on one set of 1000 rows, even if it would be inaccurate on a different 1000 rows.
The larger the validation set, the less randomness (aka "noise") there is in our measure of model quality.
In cross-validation, we run our modeling process on different subsets of the data to get multiple measures of model quality.
In Experiment 1, we use the first fold (20%) as a validation (or holdout) set and everything else as training data. We repeat this process, using every fold once as the holdout set.
Putting this together, 100% of the data is used as holdout at some point, and we end up with a measure of model quality that is based on all of the rows in the dataset.
Cross-validation gives a more accurate measure of model quality. However, it can take longer to run.
For small datasets, you should run cross-validation. But for larger datasets, a single validation set is sufficient.
There's no simple threshold for what constitutes a large vs. small dataset. But if your model takes a couple minutes or less to run, it's probably worth switching to cross-validation. Or you can run cross-validation and see if the scores for each experiment seem close.
- Setup
- Define a Pipeline
- Obtain the Cross-validation Scores
- Combine them as a Function
- Evalute the Model Performance
- Find the best Parameter Value
# load data
import pandas as pd
X_full = pd.read_csv("../input/train.csv", index_col="Id")# remove rows with missing target
X_full.dropna(axis=0, subset=["SalePrice"], inplace=True)# separate target (y) from features (X)
y = X_full["SalePrice"]
X_full.drop(["SalePrice"], axis=1, inplace=True)# select numeric columns only
numeric_cols = [
cname for cname in X_full.columns if X_full[cname].dtype in ["int64", "float64"]
]
X = X_full[numeric_cols].copy()
X_test = test_data[numeric_cols].copy()It's difficult to do cross-validation without pipelines.
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestRegressor
my_pipeline = Pipeline(
steps=[
("preprocessor", SimpleImputer()),
("model", RandomForestRegressor(n_estimators=50, random_state=0)),
]
)with the cross_val_score() function
from sklearn.model_selection import cross_val_score
scores = -1 * cross_val_score(my_pipeline, X, y, cv=5, scoring="neg_mean_absolute_error")
scores
>>> [301628 303164 287298 236061 260383]
# take the average score across experiments
scores.mean()
>>> 277707- The
cvparameter sets the number of folds. - The
scoringparameter chooses a measure of model quality to report. The docs for scikit-learn show a list of options. - It is a little surprising that we specify negative MAE. Scikit-learn has a convention where all metrics are defined so a high number is better. Using negatives here allows them to be consistent with that convention. So multiply this score by -1.
# get validation scores based on different numbers of estimators (trees)
def get_score(n_estimators):
"""return the average MAE over 3 CV folds of random forest model."""
my_pipeline = Pipeline(
steps=[
("preprocessor", SimpleImputer()),
("model", RandomForestRegressor(n_estimators=n_estimators, random_state=0)),
]
)
scores = -1 * cross_val_score(
my_pipeline, X, y, cv=3, scoring="neg_mean_absolute_error"
)
return scores.mean()Corresponding to some different values.
# for example 50, 100, 150, ..., 300, 350, 400
results = {i: get_score(i) for i in range(50, 450, 50)}import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(results.keys(), results.values())
plt.show()If you'd like to learn more about hyperparameter optimization, you're encouraged to start with grid search, which is a straightforward method for determining the best combination of parameters for a machine learning model. Thankfully, scikit-learn also contains a built-in function GridSearchCV() that can make your grid search code very efficient!
The most accurate modeling technique for structured data. #
Ensemble methods combine the predictions of several models and achieve better performance, like Random Forest method.
It is a ensemble method that goes through cycles to iteratively add models into an ensemble. Steps are:
- It begins by initializing the ensemble with a naive model, even if its predictions are wildly inaccurate.
- Then it uses the all models in the current ensemble to generate predictions for each observation in the dataset.
- These predictions are used to calculate a loss function (like mean squared error, for instance).
- Then, it uses the loss function to fit a new model that will be added to the ensemble and reduce the loss. (It uses Gradient Descent on the loss function to determine the parameters in this new model.)
- Then it adds the new model to ensemble.
- ... repeat steps 2-5!
XGBoost stands for extreme gradient boosting, which is an implementation of gradient boosting with several additional features focused on performance and speed. (Scikit-learn has another version of gradient boosting, but XGBoost has some technical advantages.)
- Setup
XGBRegressor- Make Predictions
- Parameter Tuning
# load data
import pandas as pd
X_full = pd.read_csv("../input/train.csv", index_col="Id")
X_test_full = pd.read_csv("../input/test.csv", index_col="Id")# remove rows with missing target
X_full.dropna(axis=0, subset=["SalePrice"], inplace=True)# separate target (y) from features (X)
y = X_full["SalePrice"]
X = X_full.drop(["SalePrice"], axis=1, inplace=True)# break off validation set from training data
from sklearn.model_selection import train_test_split
X_train_full, X_valid_full, y_train, y_valid = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=0
)# select categorical columns with relatively low cardinality (convenient but arbitrary)
# cardinality means the number of unique values in a column
low_cardinality_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].nunique() < 10 and X_train_full[cname].dtype == "object"
]
# select numeric columns
numeric_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].dtype in ["int64", "float64"]
]
# keep selected columns only
my_cols = low_cardinality_cols + numeric_cols
X_train = X_train_full[my_cols].copy()
X_valid = X_valid_full[my_cols].copy()
X_test = X_test_full[my_cols].copy()# one-hot encode the data
# to shorten the code, we use pandas
X_train = pd.get_dummies(X_train)
X_valid = pd.get_dummies(X_valid)
X_test = pd.get_dummies(X_test)
X_train, X_valid = X_train.align(X_valid, join="left", axis=1)
X_train, X_test = X_train.align(X_test, join="left", axis=1)The scikit-learn API for XGBoost.
# define model
from xgboost import XGBRegressor
my_model = XGBRegressor()
# fit model
my_model.fit(X_train, y_train)# make predictions
predictions = my_model.predict(X_valid)
# evaluate the model
from sklearn.metrics import mean_absolute_error
mean_absolute_error(predictions, y_valid)my_model = XGBRegressor(n_estimators=200, learning_rate=0.05, n_jobs=4)
my_model.fit(
X_train,
y_train,
early_stopping_rounds=5,
eval_set=[(X_valid, y_valid)],
verbose=False,
)n_estimators
- It specifies how many times to go through the modeling cycle. It is equal to the number of models that we include in the ensemble.
- Too low a value causes underfitting.
- Too high a value causes overfitting.
- Typical values range from 100-1000, though this depends a lot on the
learning_rateparameter.
early_stopping_rounds
- It offers a way to automatically find the ideal value for
n_estimators. - Early stopping causes the model to stop iterating when the validation score stops improving, even if we aren't at the hard stop for
n_estimators. - Since random chance sometimes causes a single round where validation scores don't improve, you need to specify a number for how many rounds of straight deterioration to allow before stopping.
- Setting
early_stopping_rounds=5is a reasonable choice. - When using
early_stopping_rounds, you also need to set aside some data for calculating the validation scores. This is done by setting theeval_setparameter.
learning_rate
- Instead of getting predictions by simply adding up the predictions from each component model, we can multiply the predictions from each model by a small number (known as the learning rate) before adding them in. This means each tree we add to the ensemble helps us less. So, we can set a higher value for
n_estimatorswithout overfitting. - If we use early stopping, the appropriate number of trees will be determined automatically.
- In general, a small learning rate and large number of estimators will yield more accurate XGBoost models, though it will also take the model longer to train since it does more iterations through the cycle. As default, XGBoost sets
learning_rate=0.1.
n_jobs
- You can use parallelism to build your models faster. It's common to set the parameter
n_jobsequal to the number of cores on your machine. - On smaller datasets, this won't help. But, it's useful in large datasets where you would otherwise spend a long time waiting during the fit command.
Find and fix this problem that ruins your model in subtle ways. #
Data leakage (or leakage) happens when your training data contains information about the target, but similar data will not be available when the model is used for prediction.
This causes a model to look accurate until you start making decisions with the model, and then the model becomes very inaccurate.
There are two main types of leakage: target leakage and train-test contamination.
It occurs when your predictors include data becomes available after predictions.
Example: People take antibiotic medicines after getting pneumonia in order to recover.
- The data shows a strong relationship between those columns.
- But
took_antibiotic_medicineis frequently changed after the value forgot_pneumoniais determined. - The model would see that anyone who has a value of
Falsefortook_antibiotic_medicinedidn't have pneumonia. - Since validation data comes from the same source as training data, the pattern will repeat itself in validation, and the model will have great validation (or cross-validation) scores.
Prevent
Any variable updated (or created) after the target value is realized should be excluded.
It occurs when the validation data affects the preprocessing behavior.
Example: Imagine you run preprocessing (like fitting an imputer for missing values) before calling train_test_split().
This problem becomes even more dangerous when you do more complex feature engineering.
Prevent
- If your validation is based on a simple train-test split, exclude the validation data from any type of fitting, including the fitting of preprocessing steps.
- This is easier if you use scikit-learn pipelines.
- When using cross-validation, it's even more critical that you do your preprocessing inside the pipeline!
Build a model to predict how many shoelaces NIKE needs each month.
The most important features in the model are
- The current month
- Advertising expenditures in the previous month
- Various macroeconomic features (like the unemployment rate) as of the beginning of the current month
- The amount of leather they ended up using in the current month
The results show the model is almost perfectly accurate if you include the feature about how much leather they used because the amount of leather they use is a perfect indicator of how many shoes they produce.
Is the leather used feature constitutes a source of data leakage?
Solution
- It depends on details of how data is collected (which is common when thinking about leakage).
- Would you at the beginning of the month decide how much leather will be used that month? If so, this is ok. But if that is determined during the month, you would not have access to it when you make the prediction.
You could use the amount of leather they ordered (rather than the amount they actually used) leading up to a given month as a predictor in your shoelace model.
Is this constitutes a source of data leakage?
Solution
- This could be fine, but it depends on whether they order shoelaces first or leather first.
- If they order shoelaces first, you won't know how much leather they've ordered when you predict their shoelace needs.
Build a model to predict the price of a new cryptocurrency one day ahead.
The most important features in the model are
- Current price of the currency
- Amount of the currency sold in the last 24 hours
- Change in the currency price in the last 24 hours
- Change in the currency price in the last 1 hour
- Number of new tweets in the last 24 hours that mention the currency
The value of the cryptocurrency in dollars has fluctuated up and down by over 100$ in the last year, and yet the model′s average error is less than 1$.
Do you invest based on this model?
Solution
- There is no source of leakage here. These features should be available at the moment you want to make a predition, and they're unlikely to be changed in the training data after the prediction target is determined.
- But, this model's accuracy could be misleading if you aren't careful.
- If the price moves gradually, today's price will be an accurate predictor of tomorrow's price, but it may not tell you whether it's a good time to invest.
- A better prediction target would be the change in price (up or down and by how much) over the next day.
Build a model to predict housing prices.
The most important features in the model are
- Size of the house (in square meters)
- Average sales price of homes in the same neighborhood
- Latitude and longitude of the house
- Whether the house has a basement
Which of the features is most likely to be a source of leakage?
Solution:
- Average sales price of homes in the same neighborhood is the source of target leakage.
- We don't know the rules for when this is updated.
- If the field is updated in the raw data after a home was sold, and the home's sale is used to calculate the average, this constitutes a case of target leakage.
- At an extreme, if only one home is sold in the neighborhood, and it is the home we are trying to predict, then the average will be exactly equal to the value we are trying to predict.
- In general, for neighborhoods with few sales, the model will perform very well on the training data. But when you apply the model, the home you are predicting won't have been sold yet, so this feature won't work the same as it did in the training data.
- Other features don't change, and will be available at the time we want to make a prediction.
Build a model to predict which applications were accepted.
# load data
import pandas as pd
data = pd.read_csv(
"../input/aer-credit-card-data/AER_credit_card_data.csv",
true_values=["yes"],
false_values=["no"],
)# separate target (y) from features (X)
y = data["card"]
X = data.drop(["card"], axis=1)Since this is a small dataset, we will use cross-validation to ensure accurate measures of model quality.
# since there is no preprocessing, we don't need a pipeline (used anyway as best practice!)
from sklearn.pipeline import make_pipeline
from sklearn.ensemble import RandomForestClassifier
my_pipeline = make_pipeline(RandomForestClassifier(n_estimators=100))
# evalutae model
from sklearn.model_selection import cross_val_score
cv_scores = cross_val_score(my_pipeline, X, y, cv=5, scoring="accuracy")
cv_scores.mean()
>>> 0.981043With experience, you'll find that it's very rare to find models that are accurate 98% of the time. It happens, but it's uncommon enough that we should inspect the data more closely for target leakage.
Summary of the data:
- card: 1 if credit card application accepted, 0 if not
- reports: Number of major derogatory reports
- age: Age n years plus twelfths of a year
- income: Yearly income (divided by 10,000)
- share: Ratio of monthly credit card expenditure to yearly income
- expenditure: Average monthly credit card expenditure
- owner: 1 if owns home, 0 if rents
- selfempl: 1 if self-employed, 0 if not
- dependents: 1 + number of dependents
- months: Months living at current address
- majorcards: Number of major credit cards held
- active: Number of active credit accounts
A few variables look suspicious. For example, does expenditure mean expenditure on this card or on cards used before appying?
# fraction of those who received a card and had no expenditures
(X["expenditure"][y] == 0).mean()
>>> 0.02# fraction of those who did not receive a card and had no expenditures
(X["expenditure"][~y] == 0).mean()
>>> 1.00As shown above, everyone who did not receive a card had no expenditures, while only 2% of those who received a card had no expenditures. This seems to be a case of target leakage, where expenditures probably means expenditures on the card they applied for.
Since share is partially determined by expenditure, it should be excluded too.
The variables active and majorcards are a little less clear, but from the description, they sound concerning.
# drop leaky features from dataset
potential_leaks = ["expenditure", "share", "active", "majorcards"]
X2 = X.drop(potential_leaks, axis=1)
# evaluate the model, with leaky predictors removed
cv_scores = cross_val_score(my_pipeline, X2, y, cv=5, scoring="accuracy")
cv_scores.mean()
>>> 0.831679Discover the most effective way to improve your models.
In this micro-course, you will learn a practical approach to feature engineering:
- Develop a baseline model for comparing performance on models with more features
- Encode categorical features so the model can make better use of the information
- Generate new features to provide more information for the model
- Select features to reduce overfitting and increase prediction speed
Building a baseline model as a starting point for feature engineering. #
We'll apply the techniques using data from Kickstarter projects. What we can do here is predict if a Kickstarter project will succeed. We get the outcome from the state column. To predict the outcome we can use features such as category, currency, funding goal, country, and when it was launched.
import pandas as pd
ks = pd.read_csv(
"../input/kickstarter-projects/ks-projects-201801.csv",
parse_dates=["deadline", "launched"],
)We'll convert the column into something can be used as targets in a model.
# look at project states
ks["state"].unique()
# how many records of each?
ks.groupby("state")["ID"].count()# drop live projects
ks = ks.query('state != "live"')
# add outcome column, "successful" = 1, others are 0
ks = ks.assign(outcome=(ks["state"] == "successful").astype(int))We can convert timestamp features into numeric catagorical features we can use in a model. If we loaded a column as timestamp data, we access date and time values through the .dt attribute on that column.
ks = ks.assign(
hour=ks["launched"].dt.hour,
day=ks["launched"].dt.day,
month=ks["launched"].dt.month,
year=ks["launched"].dt.year,
)Now for the categorical variables (category, currency, and country), we'll need to convert them into integers so our model can use the data. For this we'll use scikit-learn's LabelEncoder.
- One-Hot encoding in high cardinality features will create an extremely sparse matrix, and will make your model run very slow, so in general you want to avoid one-hot encoding features with many levels. In addition, LightGBM models work with label encoded features, so you don't actually need to one-hot encode the categorical features.
# choose catagorical features
cat_features = ["category", "currency", "country"]# create the encoder
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
# encode catagorical features
encoded = ks[cat_features].apply(le.fit_transform)# join `ks` and `encoded`. since they have the same index, we can easily join them
data = ks[["goal", "hour", "day", "month", "year", "outcome"]].join(encoded)This is a timeseries dataset. Is there any special consideration when creating train/test splits for timeseries?
- Since our model is meant to predict events in the future, we must also validate the model on events in the future. If the data is mixed up between the training and test sets, then future data will leak in to the model and our validation results will overestimate the performance on new data.
We'll use a fairly simple approach and split the data using slices. We'll use 10% of the data as a validation set, 10% for testing, and the other 80% for training.
# split data
valid_fraction = 0.1
valid_size = int(len(ks) * valid_fraction)
train = data[: -2 * valid_size]
valid = data[-2 * valid_size : -valid_size]
test = data[-valid_size:]In general we want to be careful that each data set has the same proportion of target classes. A good way to do this automatically is with sklearn.model_selection.StratifiedShuffleSplit.
# test of target classes distribution
for each in [train, valid, test]:
# because outcome is 1 or 0
print(each["outcome"].mean())For this course we'll be using a LightGBM model. This is a tree-based model that typically provides the best performance, even compared to XGBoost. It's also relatively fast to train. We won't do hyperparameter optimization because that isn't the goal of this course. So, our models won't be the absolute best performance you can get. But you'll still see model performance improve as we do feature engineering.
# define features
feature_cols = train.columns.drop("outcome")
# define train and valid datasets
import lightgbm as lgb
train_data = lgb.Dataset(train[feature_cols], label=train["outcome"])
valid_data = lgb.Dataset(valid[feature_cols], label=valid["outcome"])# fit model
param = {"num_leaves": 64, "objective": "binary", "metric": ["auc"], "seed": 7}
bst = lgb.train(
param,
train_data,
num_boost_round=1000,
valid_sets=[valid_data],
early_stopping_rounds=10,
verbose_eval=False,
)metric(s) will be used to evaluate on the evaluation set(s).aucstands for Area Under the Curve.
- If you have a validation set, you can use early stopping to find the optimal number of boosting rounds. Early stopping requires at least one set in
valid_sets. If there is more than one, it will use all of them except the training data. The model will train until the validation score stops improving. Validation score needs to improve at least everyearly_stopping_roundsto continue training. - More info about Python Quick Start, Parameters & Parameters Tuning
# make predictions
y_pred = bst.predict(test[feature_cols])# evalutae the model, based on metric(s)
from sklearn.metrics import roc_auc_score
roc_auc_score(test["outcome"], y_pred)
>>> 0.7476There are many ways to encode categorical data for modeling. Some are pretty clever. #
Now that we've built a baseline model, we are ready to improve it with some clever ways to convert categorical variables into numerical features. The most basic encodings are one-hot encoding and label encoding. Here, we'll learn count encoding, target encoding (and variations), and singular value decomposition. We'll use the categorical-encodings package to get these encodings. These encoders work like scikit-learn transformers with .fit and .transform methods. We'll compare our encodings with the baseline model from the first tutorial.
# function that splits data
def get_data_splits(data, valid_fraction=0.1):
valid_size = int(len(data) * valid_fraction)
train = data[: -valid_size * 2]
valid = data[-valid_size * 2 : -valid_size]
test = data[-valid_size:]
return train, valid, test# function that trains a model
def train_model(train, valid, test=None, feature_cols=None):
# define features
if feature_cols is None:
feature_cols = train.columns.drop("outcome")
# define train and valid datasets
import lightgbm as lgb
train_data = lgb.Dataset(train[feature_cols], label=train["outcome"])
valid_data = lgb.Dataset(valid[feature_cols], label=valid["outcome"])
# fit model
param = {"num_leaves": 64, "objective": "binary", "metric": ["auc"], "seed": 7}
bst = lgb.train(
param,
train_data,
num_boost_round=1000,
valid_sets=[valid_data],
early_stopping_rounds=10,
verbose_eval=False,
)
# make predictions
valid_pred = bst.predict(valid[feature_cols])
# evaluate the model
from sklearn.metrics import roc_auc_score
valid_score = roc_auc_score(valid["outcome"], valid_pred)
if test is not None:
test_pred = bst.predict(test[feature_cols])
test_score = roc_auc_score(test["outcome"], test_pred)
return bst, valid_score, test_score
else:
return bst, valid_score# split data
train, valid, test = get_data_splits(baseline_data)
# train a model on the baseline data
bst = train_model(train, valid)
>>> 0.7467CountEncoder replaces each categorical value with the number of times it appears in the dataset. So, rare values tend to have similar counts (with values like 1 or 2), so we can classify rare values together at prediction time. Common values with large counts are unlikely to have the same exact count as other values. So, the common/important values get their own grouping.
TargetEncoder replaces a categorical value with the average value of the target for that value of the feature. This is often blended with the target probability over the entire dataset to reduce the variance of values with few occurences. This technique uses the targets to create new features. So including the validation or test data in the target encodings would be a form of target leakage. Instead, we should learn the target encodings from the training dataset only and apply it to the other datasets.
CatBoostEncoder, similar to TargetEncoder, is based on the target probablity for a given value. However with CatBoost, for each row, the target probability is calculated only from the rows before it.
# split data
train, valid, test = get_data_splits(baseline_data)# choose catagorical features
cat_features = ["category", "currency", "country"]# create the encoder
from category_encoders import CountEncoder
ce = CountEncoder(cols=cat_features)TargetEncoder,CatBoostEncoderare the same.
# learn encoding from the training features
ce.fit(train[cat_features])TargetEncoder,CatBoostEncoderare the same, but also target must be set infit()method too. For instancete.fit(train[cat_features], train["outcome"]).
# apply encoding to the train and validation sets as new columns with a suffix
train_encoded = train.join(ce.transform(train[cat_features]).add_suffix("_ce"))
valid_encoded = valid.join(ce.transform(valid[cat_features]).add_suffix("_ce"))# fit & evaluate a model on the encoded data
bst = train_model(train_encoded, valid_encoded)We'll keep those work best. For instance CatBoostEncoder is that one.
encoded = cb.transform(baseline_data[cat_features])
for col in encoded:
baseline_data.insert(len(baseline_data.columns), col + "_cb", encoded[col])The frequently useful case where you can combine data from multiple rows into useful features. #
Creating new features from the raw data is one of the best ways to improve your model. The features you create are different for every dataset, so it takes a bit of creativity and experimentation.
# load data
import pandas as pd
ks = pd.read_csv(
"../input/kickstarter-projects/ks-projects-201801.csv",
parse_dates=["deadline", "launched"],
)
# drop live projects
ks = ks.query('state != "live"')
# add outcome column, "successful" = 1, others are 0
ks = ks.assign(outcome=(ks["state"] == "successful").astype(int))
# timestamp features
ks = ks.assign(
hour=ks["launched"].dt.hour,
day=ks["launched"].dt.day,
month=ks["launched"].dt.month,
year=ks["launched"].dt.year,
)
# choose catagorical features
cat_features = ["category", "currency", "country"]
# create the encoder
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
# encode catagorical features
encoded = ks[cat_features].apply(le.fit_transform)
# join `ks` and `encoded`
baseline_data = ks[["goal", "hour", "day", "month", "year", "outcome"]].join(encoded)One of the easiest ways to create new features is by combining categorical variables. For example, if one record has the country CA and category Music, you can create a new value CA_Music. This is a new categorical feature that can provide information about correlations between categorical variables. This type of feature is typically called an interaction.
The easiest way to iterate through the pairs of features is with itertools.combinations.
# choose catagorical features
cat_features = ["category", "currency", "country"]# create an empty DataFrame with same index
interactions = pd.DataFrame(index=ks.index)
# iterate through each pair of features & combine them
from itertools import combinations
for cname1, cname2 in combinations(cat_features, 2):
# create new cname
new_cname = "_".join([cname1, cname2])
# convert to strings and combine
new_values = ks[cname1].map(str) + "_" + ks[cname2].map(str)
# encode the interaction feature
le = LabelEncoder()
interactions[new_cname] = le.fit_transform(new_values)# join interaction features to the data
baseline_data = baseline_data.join(interactions)We'll count the number of projects launched in the preceeding week for each record. With a timeseries index, you can use .rolling to select time periods as the window. For example .rolling('7d') creates a rolling window that contains all the data in the previous 7 days. The window contains the current record, so if we want to count all the previous projects but not the current one, we'll need to subtract 1.
We'll use the .rolling method on a series with the launched column as the index. We'll create the series, using ks["launched"] as the index and ks.index as the values, then sort the times. Using a time series as the index allows us to define the rolling window size in terms of hours, days, weeks, etc.
# function takes a timestamps Series, returns another Series with the number of events in time periods
def count_past_events(series, time_window):
# create a Series with a timestamp index
series = pd.Series(series.index, index=series).sort_index()
# subtract 1 so the current event isn't counted
past_events = series.rolling(time_window).count() - 1
# adjust the index, so we can join it to the other training data
past_events.index = series.values
past_events = past_events.reindex(series.index)
return past_events# count past events, in the previous 7 days
count_7_days = count_past_events(ks["launched"], time_window="7d")# join new features to the data
baseline_data.join(count_7_days)Do projects in the same category compete for donors? We can capture this by calculating the time since the last launch project in the same category.
A handy method for performing operations within groups is to use .groupby then .transform. The .transform method takes a function then passes a series or dataframe to that function for each group. This returns a dataframe with the same indices as the original dataframe.
# function calculates the time differences (in hours)
def time_since_last_project(series):
return series.diff().dt.total_seconds() / 3600# calculate the time differences for each category
df = ks[["category", "launched"]].sort_values("launched")
timedeltas = df.groupby("category").transform(time_since_last_project)We get NaNs here for projects that are the first in their category.
timedeltas = timedeltas.fillna(timedeltas.median()).reindex(baseline_data.index)# join new features to the data
baseline_data.join(timedeltas)So far we've been using LightGBM, a tree-based model. Would these features we've generated work well for neural networks as well as tree-based models?
The features themselves will work for either model. However, numerical inputs to neural networks need to be standardized first. So, the features need to be scaled such that they have 0 mean and a standard deviation of 1. This can be done using sklearn.preprocessing.StandardScaler.
Some models work better when the features are normally distributed. Common choices for this are the log and power transformations. The log transformation won't help tree-based models because they are scale invariant. However, this should help if we had a linear model or neural network.
Other transformations include squares and other powers, exponentials, etc. These might help the model discriminate, like the kernel trick for SVMs. But again, it takes a bit of experimentation to see what works.
You can make a lot of features. Here's how to get the best set of features for your model. #
Now that we've generated hundreds or thousands of features after various encodings and feature generation, we should remove some of them to:
- Reduce the size of the model and speed up model-training and hyperparameters-optimizing, especially when building user-facing products.
- Prevent overfitting the model to the training and validation sets and improve the performance.
The simplest and fastest methods are based on univariate statistical tests. For each feature, measure how strongly the target depends on the feature using a statistical test like Ⲭ² or ANOVA.
From the sklearn.feature_selection module, SelectKBest returns the K-best features given some scoring function. For our classification problem, the module provides three different scoring functions: Ⲭ², ANOVA F-value, and the mutual information score.
- The F-value measures the linear dependency between the feature variable and the target. This means the score might underestimate the relation between a feature and the target if the relationship is nonlinear.
- The mutual information score is nonparametric and so can capture nonlinear relationships.
With SelectKBest, we define the number of features to keep, based on the score from the scoring function. Using .fit_transform we get back an array with only the selected features.
# split data
train, valid, test = get_data_splits(baseline_data)# choose features
feature_cols = baseline_data.columns.drop("outcome")
# seperate predictors (X) and target (y)
X, y = train[feature_cols], train["outcome"]# keep 5-best features
from sklearn.feature_selection import SelectKBest, f_classif
selector = SelectKBest(f_classif, k=5)
X_new = selector.fit_transform(X, y)Now we have our selected features. To drop the rejected features, we need to figure out which columns in the dataset were kept with SelectKBest. To do this, we can use .inverse_transform to get back an array with the shape of the original data. This returns a DataFrame with the same index and columns as the training set, but all the dropped columns are filled with zeros. We can find the selected columns by choosing features where the variance is non-zero.
# get back the features we've kept
selected_features = pd.DataFrame(
selector.inverse_transform(X_new), index=X.index, columns=feature_cols
)
# drop columns have values of all 0s
selected_columns = selected_features.columns[selected_features.var() != 0]With this method we can choose the best K features, but we still have to choose K ourselves. How would we find the "best" value of K?
- To find the best value of K, we can fit multiple models with increasing values of K, then choose the smallest K with validation score above some threshold or some other criteria. That is, we're keeping the best features w/o degrading the model's performance.
Univariate methods consider only one feature at a time when making a selection decision. Instead, we can make our selection using all of the features by including them in a linear model with L1 (Lasso) regularization which penalizes the absolute magnitude of the coefficients, or L2 (Ridge) regression which penalizes the square of the coefficients.
In general, feature selection with L1 regularization is more powerful the univariate tests, but it can also be very slow when you have a lot of data and a lot of features. As the strength of regularization is increased, features which are less important for predicting the target are set to 0.
From sklearn.linear_model module, you can use Lasso for regression problems, or LogisticRegression for classification. These can be used along with sklearn.feature_selection.SelectFromModel to select the non-zero coefficients. Otherwise, the code is similar to the univariate tests.
# split data
train, valid, test = get_data_splits(baseline_data)# choose features
feature_cols = baseline_data.columns.drop("outcome")
# seperate predictors (X) and target (y)
X, y = train[feature_cols], train["outcome"]from sklearn.linear_model import LogisticRegression
from sklearn.feature_selection import SelectFromModel
logistic = LogisticRegression(C=1, penalty="l1", random_state=7).fit(X, y)
selector = SelectFromModel(logistic, prefit=True)
X_new = selector.transform(X)Cis the inverse of regularization strength, which leads to some number of features being dropped. However, by setting C we aren't able to choose a certain number of features to keep. To find the regularization parameter that leaves the desired number of features, we can iterate over models with different regularization parameters from low to high and choose the one that leaves K features.prefitdetermines a model to be passed into the constructor directly or not. If True,transformmust be called directly andSelectFromModelcannot be used withcross_val_score,GridSearchCVand similar utilities that clone the estimator. Otherwise train the model usingfitand thentransformto do feature selection.
# get back the features we've kept
selected_features = pd.DataFrame(
selector.inverse_transform(X_new), index=X.index, columns=feature_cols
)
# drop columns have values of all 0s
selected_columns = selected_features.columns[selected_features.var() != 0]Since we're using a tree-based model, using another tree-based model for feature selection might produce better results. We could use something like RandomForestClassifier or ExtraTreesClassifier to find feature importances. SelectFromModel can use the feature importances to find the best features.
Use TensorFlow to take machine learning to the next level. Your new skills will amaze you.
A quick overview of how models work on images. #
Deep Learning has revolutionized computer vision and it is the core technology behind capabilities like self driving cars.
For this series, we will use tensorflow and the tensorflow implementation of keras. Tensorflow is the most popular tool for deep learning and keras is a popular api or for specifying deep learning models. This allows you to specify models with the elegant of keras while taking greater advantage of some powerful tensorflow features.
Your machine learning experience so far has probably focused on tabular data like you would work with in pandas. We will be doing deep learning with images so we will start an overview of how images are stored for machine learning.
Images are composed of pixels.
- In b/w images, some pixels are completely black, some are completely white, and some are varying shades of grey. The pixels are arranged in rows and columns so it is natural to store them in a matrix. Each value represents the darkness of the pixel in the image
- Colour images have an extra dimension. For each pixel we store how red that pixel is, how green it is, and how blue it is. It's like a stack of three matrices.
Tensor is the word for something that is like a matrix but which can have any number of dimensions.
Today's deep learning models apply something called convolutions to this type of tensor. A convolution is a small tensor that can be multiplied over little sections of the main image. Convolutions are also called filters because depending on the values in that convolutional array, you can pick out the percific patterns from the image, like horizontal or vertical lines. The numerical values in the convolution determine what pattern it detects.
You don't directly choose the numbers to go into your convolutions for deep learning... instead the deep learning technique determines what convolutions will be useful from the data (as part of model-training). But looking closely at convolutions and how they are applied to your image will improve your intuition for these models, how they work, and how to debug them when they don't work.
For example, a convolution that detected horizontal lines:
horizontal_line_conv = [[1, 1], [-1, -1]]or, a vertical line detector:
vertical_line_conv = [[1, -1], [1, -1]]While any one convolution measures only a single pattern, there are more possible convolutions that can be created with large sizes. So there are also more patterns that can be captured with large convolutions. Does this mean powerful models require extremely large convolutions? Not necessarily.
Scale up from simple building blocks to models with beyond human capabilities. #
Once we create a filter, we applied it to each part of the image and map the output. This gave us a map showing where the associative pattern shows up in the image. Different convolutions or filters capture different aspects of the original image. In practice, we won't mainly pick the numbers in our filters. We using training process like gradient descent and back propagation to automatically create filters, which means we can have a lot of filters to capture many different patterns.
We represent an image as a matrix, or a 2d tensor. Each convolution we apply to that tensor creates a new 2d tensor. We stack all those 2d tensors into a single 3d tensor. For example, we might have a 2d tensor showing where the horizontal lines are. And we stack it on top of a 2d tensor showing where the vertical lines are and keep stacking with any other tensors from other convolutions that we have. The result is a representation of the image in three dimensions. This last dimension is called the channel dimension.
We're going to apply an another set or a layer of convolution. But we won't apply them to the raw pixel intensities. Instead, we applied this layer of convolutions to that 3d tensor, we got as the output of the first layer of convolutions. Our second layer convolutions takes that map of pattern locations as input and multiplies them with 3d convolution to find more interesting patterns.
Modern networks apply a lot of these layers. Some research papers have used over 1000 layers. After enough layers and the right numbers and the compositions are filtered, we have a 3d tensor with a very rich summary of the image content.
What can you do with the summary of the image content? We'll start with something called object detection. Your model will take a photo as input, and it returns a prediction for what type of object is in the photo. Is it a dog or a kangaroo or a robot, or whatever.
Many breakthroughs in computer vision and AI have come through an object detection competition called ImageNet. Competitors receive a training set with millions of images, each image is labelled with its content coming from 1000 different possible labels.
Competitors use this training data to build a model that classifies images into those thousand options. The best models get it right with their top prediction about 80% of the time. If each model gets five predictions, the true image label is in those top five predictions about 95% of the time. This is especially impressive because there are many similar options that the model must choose between. So labelling an image correctly requires not only that, you know, it's a picture of a shark, but you have to know that it is a hammerhead shark or a tiger shark or a great white shark. So when those best models are right on our first guest 80% of the time, that's better than most people could do.
Some models using the ImageNet competition are also available as pre trained models. So, even before you learn to train a model for yourself, you can start with powerful models that classify images into one of 1000 different categories.
As a very first step programming with Keras and TensorFlow, we will start by using a pre trained model from the ImageNet competition. Once you can run these models, we'll move on to something called transfer learning, which allows you to apply the same level of power to most other problems you might be interested in.
Start writing code using TensorFlow and Keras. #
As the first step with Keras and TensorFlow, we will use a pre trained deep learning model to classify what's in a photo. Pre trained models are saved on kaggle, you can attach them to your kernel workspace, the same way you would attach a data set.
We will work with dog pictures and try to tell each dog's breed from its picture that data is in this directory. To keep things simple, we'll make prediction with just a few of those images.
# load data
from os.path import join
image_dir = "../input/dog-breed-identification/train/"
img_paths = [
join(image_dir, filename)
for filename in [
"0c8fe33bd89646b678f6b2891df8a1c6.jpg",
"0c3b282ecbed1ca9eb17de4cb1b6e326.jpg",
"04fb4d719e9fe2b6ffe32d9ae7be8a22.jpg",
"0e79be614f12deb4f7cae18614b7391b.jpg",
]
]We'll use a type of model called the ResNet50 model (in the excercise, we'll use VGG16). But first, we need to do a little bit of pre processing to go from the image file path to something we can run through our model.
# function to read and prep images for modeling
image_size = 224
def read_and_prep_images(img_paths, img_height=image_size, img_width=image_size):
# load images
from tensorflow.keras.preprocessing.image import load_img
imgs = [
load_img(img_path, target_size=(img_height, img_width))
for img_path in img_paths
]
# convert each image to an array
import numpy as np
from tensorflow.keras.preprocessing.image import img_to_array
img_array = np.array([img_to_array(img) for img in imgs])
# preprocess on images' array
from tensorflow.keras.applications.resnet50 import preprocess_input
output = preprocess_input(img_array)
return output- The
target_sizeargument specifies the size or pixel resolution we want the images to be in when we model with them. The model will use was trained with 224 by 224 resolution images. So, we'll make them have the same resolution here. - The
img_to_arrayfunction creates the 3d tensor for each image, combining multiple images cause us to stack those in a new dimension, so we end up with a four dimensional tensor or array. - The
pre_process_inputfunction does some arithmetic on the pixel values, specifically, dividing the values in the input, so they're all between minus one and one. This was done when the model was first built, so we have to do it again here to be consistent.
# specify the model
from tensorflow.keras.applications import ResNet50
# load a pre-trained model's weights (values in the convolution filters)
my_model = ResNet50(
weights="../input/resnet50/resnet50_weights_tf_dim_ordering_tf_kernels.h5"
)# preprocess the data
test_data = read_and_prep_images(img_paths)# make predictions
preds = my_model.predict(test_data)We have predictions about what's in each image. We had four photographs and our model gave 1000 probabilities for each photo. It's convenient to focus on the probabilities for what the model thinks is in the image rather than what all the things it says are not in the image.
# function extracts the highest probabilities for each image
def decode_predictions(
preds, top=5, class_list_path="../input/resnet50/imagenet_class_index.json"
):
"""Decodes the prediction of an ImageNet model"""
if len(preds.shape) != 2 or preds.shape[1] != 1000:
raise ValueError(
"`decode_predictions` expects a batch of predictions"
"(i.e. a 2D array of shape (samples, 1000)). "
"Found array with shape: " + str(preds.shape)
)
import json
class_index = json.load(open(class_list_path))
results = []
for pred in preds:
top_indices = pred.argsort()[-top:][::-1]
result = [tuple(class_index[str(i)]) + (pred[i],) for i in top_indices]
result.sort(key=lambda x: x[2], reverse=True)
results.append(result)
return results# extract the highest probabilities
most_likely_labels = decode_predictions(
preds, top=3, class_list_path="../input/resnet50/imagenet_class_index.json"
)
# display
from IPython.display import Image, display
for i, img_path in enumerate(img_paths):
display(Image(img_path))
print(most_likely_labels[i])This may still feel like magic right now, but it will come familiar as we play with a couple of examples. Then we'll be ready for transfer learning and quickly apply these models to new applications.
A powerful technique to build highly accurate models even with limited data. #
Using a pre trained model to make predictions, gives you great result if you want to classify images into the categories used by the original models. But what if you have a new use case, and you don't categorise images in exactly the same way as the categories for the pre trained model?
For example, I might want a model that can tell if a photo was taken in an urban area, or a rural area. My pre trained model doesn't classify images into those two specific categories. We could build a new model from scratch for this specific purpose, but to get good results, we need thousands of photos with labels for which are urban, and which are rural. Something called transfer learning, will give us good results with far less data. Transfer learning takes what a model learned while solving one problem, and applies it to a new application.
Early layers of a deep learning model identify simple shapes. Later layers identify more complex visual patterns, and the very last layer makes predictions. So most layers from a pre trained model are useful in new applications because most computer vision problems involve similar low level visual patterns. So we'll reuse most of the pre trained ResNet model and just replace that final layer that was used to make predictions.
After removing the prediction layer, the last layer of what's left has information about our photo content stored as a series of numbers in a tensor. It should be a one dimensional tensor, which is also called a vector. The vector can be shown as a series of dots. Each dot is called a node and represents a feature.
We want to classify the image into two categories, urban and rural. So after the last layer of the pre trained model, we add a new layer with two nodes, one node to capture how urban the photo is and another to capture how rural it is. In theory, any node (feature) in the last layer before prediction layer might inform the predictions. So, urban measurment can depend on all the nodes in this layer. When this happens, we describe the last layer as being a dense layer.
# define a sequential model, which is a sequence of layers
from tensorflow.keras.models import Sequential
model = Sequential()# use a pre trained ResNet50 model without prediction layer
from tensorflow.keras.applications import ResNet50
model.add(
ResNet50(
include_top=False,
weights="../input/resnet50/resnet50_weights_tf_dim_ordering_tf_kernels_notop.h5",
pooling="avg",
)
)include_top=Falseis how we specify that we want to exclude the prediction layer, that makes predictions into the thousands of categories used in the ImageNet competition.- We'll also use a file that doesn't include the weights for that last layer (
..notop.h5). pooling='avg'says that if we had extra channels in our tensor at the end of this step, we want to collapse them into a 1d tensor by taking an average across channels.
# specify the number of prediction classes
num_classes = 2
# add a Dense layer to make predictions
from tensorflow.keras.layers import Dense
model.add(Dense(num_classes, activation="softmax"))- After making predictions, we'll get a score for each category. We'll apply the
softmaxfunction to transform the scores into probabilities.
We'll tell TensorFlow not to train the first layer, which is the ResNet50 model, because that's the model that was already pre trained with the ImageNet data.
# tell TF not to train first layer (ResNet) model
model.layers[0].trainable = FalseIn general, the compile command tells TensorFlow how to update the relationships in the dense connections, when we're doing the training with our data. We'll give a more complete explanation of the underlying theory later.
from tensorflow.keras.losses import categorical_crossentropy
model.compile(loss="categorical_crossentropy", optimizer="sgd", metrics=["accuracy"])lossdetermines what goal we optimize. We used a measure of loss (inaccuracy) calledcategorical_crossentropy, another term for log loss.optimizerdetermines how we minimise the loss function. We used an algorithm called stochastic gradient descent (sgd).metricsdetermines what we print out while the model is being built.accuracyis what fraction of our predictions were correct (FC). This is easier to interpret than categorical cross entropy scores.
The compiling step doesn't change the values in any convolutions, because the model has not even received an argument with data yet. So, it runs so quickly.
Our raw data is broken into a directory of training data, and a directory of validation data. Within each of those, we have one sub directory for the urban pictures, and another for the world pictures. Keras provides a great tool for working with images grouped into directories by their label. This is the ImageDataGenerator. The It is especially valuable when working with large datasets, because we don't need to hold the whole dataset in memory at once.
There's two steps to using ImageDataGenerator. First, we create the generator object in the abstract. We'll tell it that we want to apply the ResNet preprocessing function (preprocess_input) every time it reads an image, which is used when ResNet model was created.
# define data generator
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.applications.resnet50 import preprocess_input
data_generator = ImageDataGenerator(preprocessing_function=preprocess_input)
image_size = 224Then we use the flow_from_directory command. We tell it what directory that data is in, what size image we want, how many images to read in at a time, and we tell it we're classifying data into different categories.
# generate train data
train_generator = data_generator.flow_from_directory(
directory="../input/urban-and-rural-photos/rural_and_urban_photos/train",
target_size=(image_size, image_size),
batch_size=24,
class_mode="categorical",
)# generate validation data
validation_generator = data_generator.flow_from_directory(
directory="../input/urban-and-rural-photos/rural_and_urban_photos/val",
target_size=(image_size, image_size),
batch_size=20,
class_mode="categorical",
)# fit model
model.fit_generator(
train_generator,
steps_per_epoch=3,
validation_data=validation_generator,
validation_steps=1,
)train_generatorreads 24 images at a time (batch_size=24), and we have 72 images. So we'll fit model through 3steps_per_epoch.validation_generatorreads 20 images at a time (batch_size=20), and we have 20 images of validation data. So we can use just 1validation_step.
As the model training is running, we'll see progress updates showing with our loss function and the accuracy. It updates the connections in the dense layer that is the models impression of what makes an urban photo and what makes a rural photo, and it makes us update in 3 steps. When it's done, it got 73% of the train data right. Then it examines the validation data, which gets 85% of those right, 17 out of 20.
- The printed validation accuracy can be meaningfully better than the training accuracy at this stage. This can be puzzling at first. It occurs because the training accuracy was calculated at multiple points as the network was improving (the numbers in the convolutions were being updated to make the model more accurate). The network was inaccurate when the model saw the first training images, since the weights hadn't been trained/improved much yet. Those first training results were averaged into the measure above. The validation loss and accuracy measures were calculated after the model had gone through all the data. So the network had been fully trained when these scores were calculated.
We trained on 72 photos. You could easily take that many photos on your phone, upload them to kaggle and build a very accurate model to distinguish almost anything you care about. This is incredibly COOL!
Learn a simple trick that effectively increases amount of data available for model training. #
What about one of those urban images? If we flip the image horizontally, taking the photos mirror image, it would still look like an urban scene. So we can train our model with both images. At some point, we may have so many raw images that we don't need this, but it's usually valuable, even with hundreds of thousands of images.
We do these kind of triks with some arguments in ImageDataGenerator function. These techniques are listed in the documentation, which you can see in kernels by using ImageDataGenerator? command.
# define data generator with augmentation
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.applications.resnet50 import preprocess_input
data_generator_with_aug = ImageDataGenerator(
preprocessing_function=preprocess_input,
horizontal_flip=True,
width_shift_range=0.2,
height_shift_range=0.2,
)
image_size = 224- If we set
horizontal_flip=True, theImageDataGeneratorwill randomly decide whether or not to flip an image, every time it sends the image to the model for training. We need to judge whether this makes sense on a case by case basis. For instance, the mirror image of stop sign shouldn't be classified the same as the original image. - We could slightly crop the photo, effectively shifting it slightly horizontally or vertically, and I'll add commands to randomly shift part of the image that we train on 20% further to either side, and 20% further up or down. I do that by setting
width_shift_range, andheight_shift_rangeto 0.2.
# generate train data
train_generator = data_generator_with_aug.flow_from_directory(
directory="../input/urban-and-rural-photos/rural_and_urban_photos/train",
target_size=(image_size, image_size),
batch_size=24,
class_mode="categorical",
)We want to do data augmentation when fitting the model for the reasons, including a reduction in overfitting, by giving us more data to work with. But we don't want to change how we test the model. So the validation generator will use an ImageDataGenerator without augmentation or manipulation. That allows a straightforward comparison between different training procedures (e.g. training with augmentation and without it).
# define data generator w/o augmentation
data_generator_no_aug = ImageDataGenerator(preprocessing_function=preprocess_input)# generate validation data
validation_generator = data_generator_no_aug.flow_from_directory(
directory="../input/urban-and-rural-photos/rural_and_urban_photos/val",
target_size=(image_size, image_size),
batch_size=20,
class_mode="categorical",
)Now we're ready to fit the model.
# fit model
model.fit_generator(
train_generator,
epochs=2,
steps_per_epoch=3,
validation_data=validation_generator,
validation_steps=1,
)epochs=2means it goes through each raw image file, two times, since we get different versions of the images, each time we see them. Data augmentation allows us to use more epochs before we start overfitting, and seeing validation scores get worse.
Validation scores are more reliable on bigger validation datasets. In this particular case, our validation data is so small that there's a little bit of noise or luck in a score from any given model.
How Stochastic Gradient Descent and Back-Propagation train your deep learning model. #
We will loop back to some technical details and understand how we use stochastic gradient descent and back-propagation to set the numeric values in both dense and convolutional layers, as well as how arguments like batch size and number of epochs affects this model training process.
To simplify the explanation, we'll start with an example using tabular data, the type of data we might manipulate with pandas or SQL database. Imagine we want to assess someone's risk of developing diabetes in the next year. We have data about their age, weight and a blood sugar measure. We could draw a layer with one node for each variable. Since this layer represents raw data, we call this the input layer. Let's connect these directly to the prediction layer. Each person either develops diabetes in the next year or they don't. So there are two possible outcomes, and that's two nodes in a prediction or output layer. We'll connect these as a dense layer, where each input is connected to each output. Each connection will have a number associated with it. The numbers are called weights.
Let's consider a single person. We can calculate the values in the next layer using a process called forward propagation. For each node, we multiply the weight of connections by the value at the node that connection comes from, similar to the calculations we did with convolutions. Then we sum those to get a score. For the predictions, the softmax function converts those scores into probabilities.
When working with tabular data, it's common to have many dense layers between the input and the output. The same way we had many layers of convolutions when working with images. The layers between the input and output are called hidden layers. The forward propagation process remains the same, filling in the values from left to right.
In practice, we'll apply some nonlinear function at each node in the hidden layers. Including a nonlinear function helps the model capture both nonlinearities and interaction impacts between the variables better. The most common function to apply is the ReLU or rectified linear activation function.
Though, we covered forward propagation to make predictions, we want to understand where the weights on the connections come from, because changing the weights will change the predictions. So good weights are the key to good predictions.
The loss function measures how good a models predictions are. The loss functions argument are the actual values of the target that we want to predict. If the predicted values are close to the actual values of the target, the loss function will give a low value. the numeric value of the loss function will change if we change the models weights. This is the key to our model optimization procedure, (stochastic) gradient descent.
We'll use stochastic gradient descent to set the weights that minimise our loss function. The weights affect our predictions and thus they affect the loss function. We could plot the loss function against those weights. In this plot, if we had a model with only two weights, we have the loss function on the Z axis, and weights on the other axes (3D contour plot). We want to find the low point on that surface. How would we do it? We look at data and see which way we can change the weights to get a little lower loss function and we change the weights slightly in that direction. Then we repeat this to improve the weight slightly again. This is basically how gradient descent works.
In gradient descent, we generally don't use all of our data to calculate each step. Doing so would require a lot of calculations, so it would be slow. Instead, we look at some of the data at a time. The batch size is the number of rows of the tablet data, or the number of images, we use to calculate each step. We take one small batch, and then the next until we've used all of our data. One time through the data is called an epoch. We incrementally improve weights from multiple epochs. So each image or data point would be used to improve weight more than once.
Back propagation is the process by which we find out which way to change the weights at each step of gradient descent to improve the loss function.
We use forward propagation to go from input data to values in the hidden layer, and then to predictions. For each person in our training data, if we predicted the right output, our loss function would take a low value with little ability to improve further. If we predicted the wrong output, we would have a lot of room to improve. But how do we change the weights to improve? We'll go back one layer at a time. Consider a node feeding into the right prediction value with a positive value. The higher that weight is, the more likely we are to predict that node. That means the model that has a better loss function for this person. So we can increase the value of this weight and then improve the loss function, but it also might cause us to be very wrong for other people in the data set. Lowering the weights from this hidden node to the wrong prediction has a similar effect.
The value in the hidden node may have contributed to making our predictions either more or less accurate. That is, raising or lowering the value in that node might improve our predictive probabilities. We can change the value in that node by changing the weight that feed into it. If we had many layers, we would continually iterate back from right to left until we got to the leftmost set of weights.
The size of weight changes is determined by something called the learning rate. Low learning rates mean that your model may take a long time training before it gets accurate. High learning rates mean your model may take huge steps around in that field, jumping over the best weights and never getting very accurate. We can use the argument optimizer="adam" in fit (or fit_generator) function that automatically figures out the best learning rate throughout the gradient descent process.
While we've demonstrated this with dense layers, it works the same with convolutional layers.
Build models without transfer learning. Especially important for uncommon image types. #
The deep learning models you've built so far have used transfer learning. Transfer learning is great because it allows you to quickly build accurate models, even with relatively little data. But transfer learning isn't necessary on large data sets. And it only works when your use case has similar visual patterns to the data used in the pre trained model. So, if you want to be versatile with deep learning, you'll need to learn to build models from scratch.
Now, we'll use the MNIST data set, which contains handwritten digits. In the exercise, we'll build a model to identify different types of clothing from low resolution images using a data set called MNIST for fashion. Both data sets contain 28 by 28 pixel images. We can load them from a CSV file containing all the data for all the images. This is the the first time we load all the image data from a single file, rather than using ImageDataGenerator to load many separate image files.
# load data
import pandas as pd
raw_data = pd.read_csv("../input/digit-recognizer/train.csv")To store all the images in a single CSV, each image is represented as a row of the CSV. The first column has the label. The rest of the columns represent pixel intensities. The pixels are represented in the CSV as a row of 784 pixel intensities, even though the image is started as a two dimensional grid, that's 28 by 28. We'll use a data_prep function to extract labels and reshape the pixel intensity data back to a 28 by 28 grid, before applying our model.
# specify the images' dimentions
img_rows, img_cols = 28, 28
# specify the number of prediction classes
num_classes = 10import numpy as np
from tensorflow.keras.utils import to_categorical
def data_prep(raw):
# one-hot encode the catagorical data
out_y = to_categorical(raw["label"], num_classes)
# get the number of images
num_images = raw.shape[0]
# get data as a NumPy array, except the 1st column, it's the label
x_as_array = raw.values[:, 1:]
# reshape data
x_shaped_array = x_as_array.reshape(num_images, img_rows, img_cols, 1)
# normalize data
out_x = x_shaped_array / 255
return out_x, out_y- Keras has a
to_categoricalfunction. We supply the target variablerow["label"], as well as the number of different classesnum_classes. It returns a one-hot encoded version of the target. So it represents the 10 possible values of the target with 10 separate binary columns. This gives us the target in a format that Keras expects it. - We'll reshape this into a 4D array which is indexed by the image number, row number, column number, and channel. The images were grayscale rather than colour, so, there's only one channel.
- We divide the values by 255 so all the data is between zero and one. This improves optimization with a default parameters for the
"adam"optimizer.
# get an array of predictors and an array of the target
x, y = data_prep(raw_data)xis the data holding the images.yis the data with the class labels to be predicted.
# define a sequential model
from tensorflow.keras.models import Sequential
model = Sequential()# add 2D convolutional first layer
from tensorflow.keras.layers import Conv2D
model.add(
Conv2D(
20, kernel_size=(3, 3), activation="relu", input_shape=(img_rows, img_cols, 1)
)
)- For each convolution layer, we need to specify the number of convolutions (filters) to include in that layer, the
kernel_size(size of the filters), and theactivationfunction. For the very first layer only, we need to specify the shape of each image (rows, cols, channels).
# add more layers
model.add(Conv2D(20, kernel_size=(3, 3), activation="relu"))# convert the output of previous layers into a 1D representation
from tensorflow.keras.layers import Flatten
model.add(Flatten())# for better performance, add a Dense layer in between the Flatten-prediction layers
from tensorflow.keras.layers import Dense
model.add(Dense(100, activation="relu"))- For each dense layer, we need to specify the number of neurons and the
activationfunction.
# add a Dense prediction layer
model.add(Dense(num_classes, activation="softmax"))from tensorflow.keras.losses import categorical_crossentropy
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])from sklearn.model_selection import train_test_split
model.fit(x, y, batch_size=128, epochs=2, validation_split=0.2)- Because we have the whole data loaded into arrays already, we used the
fitcommand, rather than thefit_generatorcommand that we used withImageDataGenerator. - We want to get a validation score, so we used the
validation_splitargument to say that 20% of the data should be set aside for validation.
When we run this, we can watch the accuracy go from 10% at the beginning to about 98% pretty quickly. We can experiment with a few things to improve the model slightly:
- Changing the number of convolutions in any layer
- Adding or removing layers, either adding convolution layers before the flatten, or dense layers after it
- Changing the filter or kernel size in any layer.
With increase the number of layers or the number of convolutions in a layer, we're increasing the models ability to fit the training data. We call that the model capacity that can improve a model. It could cause underfitting or overfitting; So we must use validation scores as the ultimate measure of model quality as we experiment with different model architectures.
Make your models faster and reduce overfitting. #
Stride length is the number of columns/rows we slide a covolution to an image to get some filtered output. Changing it can speed up modelling and reduce memory requirements.
For example, if we moved across two columns at a time instead of one, we would apply each convolution half as many times, and the output from this layer would be half as wide. For the same reason, moving down two rows at a time would make the output half as tall. If we always move in increments of two pixels, we would say the stride length is two. Since the resulting output was half as wide and half as tall, the representation going into the next layer ends up being only a quarter as large. This makes the model much faster. Because the larger stride reduces computational demands, it might help you try using larger models with more layers or more convolutions per layer.
# add a convolution layer with strides
model.add(Conv2D(30, kernel_size=(3, 3), strides=2, activation="relu"))There are alternative ways to achieve the same effective strides including something called max pooling. But strides are conceptually, a little cleaner than the alternatives, and there doesn't seem to be a systematic difference in model performance for most applications. For a few advanced applications like generative models, increasing stride length works much better than max pooling.
Dropout is the most popular technique to combat overfitting. With dropout we ignore randomly chosen nodes or convolutions for brief parts of training, and then we'll randomly pick other nodes to ignore for the next part of training. It prevent overfitting. It makes each convolution or node, find information that is useful for predictions in its own right, rather than allowing one node to dominate the prediction, with everything else being small tweaks on that one nodes conclusion.
# add a Dropout layer
from tensorflow.keras.layers import Dropout
model.add(Dropout(0.5))- It says that each convolution in the preceding layer should be ignored or disconnected from the subsequent layer 50% of the time during model training.
Even when we use dropout as part of model training to get better weights, all nodes and all connections are used when we make predictions.
Before we knew about dropout, researchers addressed overfitting by limiting model capacity. That meant they had fewer layers and fewer nodes or convolutions per layer. But it's common now to build very large networks that could easily overfit, and then address overfitting by adding dropout.
- Written letter recognition
- Flower Identification
- Cats vs Dogs
- 10 Monkeys
- Predict Bone Age from X-Rays
Learn SQL for working with databases, using Google BigQuery to scale to massive datasets.
Learn the workflow for handling big datasets with BigQuery and SQL. #
SQL programming is an important skill for any data scientist. In this course, we'll build our SQL skills using BigQuery, a web service that lets you apply SQL to huge datasets. To use BigQuery, the first step is to create a Client object.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()In BigQuery, each dataset is contained in a corresponding project. We'll work with a dataset of posts on Hacker News, and our hacker_news dataset is contained in the bigquery-public-data project.
# construct a reference to the "hacker_news" dataset
dataset_ref = client.dataset("hacker_news", project="bigquery-public-data")
# fetch the dataset (API request)
dataset = client.get_dataset(dataset_ref)Every dataset is just a collection of tables.
# print names of all tables
[table.table_id for table in client.list_tables(dataset)]
>>> ['comments', 'full', 'full_201510', 'stories']Similar to how we fetched a dataset, we can fetch a table.
# construct a reference to the "full" table
table_ref = dataset_ref.table("full")
# fetch the table (API request)
table = client.get_table(table_ref)The structure of a table is called its schema. We need to understand a table's schema to effectively pull out the data we want.
# print information on all the columns in the "full" table
table.schemaEach SchemaField tells us about a specific column (field):
- The name of the column
- The field type (or datatype) in the column
- The mode of the column ('NULLABLE' means that a column allows NULL values, and is the default)
- A description of the data in that column
We can use the list_rows() method to check just the first five lines of of the table to make sure this is right. (Sometimes databases have outdated descriptions, so it's good to check.) This returns a BigQuery RowIterator object that can quickly be converted to a pandas DataFrame with the to_dataframe() method.
# preview the first five lines of the table
client.list_rows(table, max_results=5).to_dataframe()# preview the first five entries in the first column of the table
client.list_rows(table, selected_fields=table.schema[:1], max_results=5).to_dataframe()More info: this link
The foundational compontents for all SQL queries. #
We'll use the OpenAQ dataset about air quality.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# construct a reference to the "openaq" dataset
dataset_ref = client.dataset("openaq", project="bigquery-public-data")
# fetch the dataset (API request)
dataset = client.get_dataset(dataset_ref)# print names of all tables
[table.table_id for table in client.list_tables(dataset)]
>>> ['global_air_quality']# construct a reference to the "global_air_quality" table
table_ref = dataset_ref.table("global_air_quality")
# fetch the table (API request)
table = client.get_table(table_ref)# preview the first five lines of the "global_air_quality" table
client.list_rows(table, max_results=5).to_dataframe()The most basic SQL query selects a single column from a single table. We do this by specify:
- the column(s) after the word
SELECT, - the table after the word
FROM, - the condition(s) after the word
WHERE.
# query to select all the items from the "city" column where the "country" column is 'US'
# in the project "bigquery-public-data", dataset "openaq", table "global_air_quality".
query = """
SELECT
city
FROM
`bigquery-public-data.openaq.global_air_quality`
WHERE
country = 'US'
"""- The argument we pass to FROM is in backticks (`), not in quotation marks (' or ").
We set up the query with the query() method.
# set up the query
query_job = client.query(query)This method allows us to specify complicated settings. For example, working with BIG datasets. BigQuery datasets can be huge. The biggest dataset currently on Kaggle is 3TB, so we can go through our 5TB-every-30-day-data-scan limit in a couple queries. We can avoid that with some tricks.
We can estimate the size of any query before running it. To see how much data a query will scan, we create a QueryJobConfig object and use the dry_run parameter.
# setup the query (estimate the size of a query without running it)
dry_run_config = bigquery.QueryJobConfig(dry_run=True)
query_job = client.query(query, job_config=dry_run_config)# print estimated cost in bytes
query_job.total_bytes_processed
>>> 386485420Or, we can create QueryJobConfig object and use the maximum_bytes_billed parameter to limit how much data we're willing to scan. The query will be cancelled if the limit was exceeded.
# setup the query (run the query if it's less than 1 GB, cancel if it exeeds)
safe_config = bigquery.QueryJobConfig(maximum_bytes_billed=10 ** 9)
query_job = client.query(query, job_config=safe_config)Then, we submit the query to the dataset (API request) and convert the results to a DataFrame.
# try to run the query, and return a pandas DataFrame (API request)
result_df = query_job.to_dataframe()Get more interesting insights directly from your SQL queries. #
We'll use the Hacker News dataset to find which comments generated the most discussion.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# construct a reference to the "hacker_news" dataset
dataset_ref = client.dataset("hacker_news", project="bigquery-public-data")
# fetch the dataset (API request)
dataset = client.get_dataset(dataset_ref)# construct a reference to the "comments" table
table_ref = dataset_ref.table("comments")
# fetch the table (API request)
table = client.get_table(table_ref)- the
parentcolumn indicates the comment that was replied to, and - the
idcolumn has the unique ID used to identify each comment
To answer more interesting questions, we group data and count things within those groups. We do that by using 3 techniques.
COUNT()returns a count of things. If you pass it the name of a column, it will return the number of entries in that column.COUNT()is an example of an aggregate function, which takes many values and returns one. (Other examples of aggregate functions includeSUM(),AVG(),MIN(), andMAX().)
GROUP BYtakes the name of one or more columns, and treats all rows with the same value in that column as a single group when you apply aggregate functions likeCOUNT().- In SQL, it doesn't make sense to use
GROUP BYwithout an aggregate function. So, if we have anyGROUP BYclause, then all variables must be passed to either aGROUP BYcommand, or an aggregation function.
- In SQL, it doesn't make sense to use
HAVINGis used in combination withGROUP BYto ignore groups that don't meet certain criteria.
# query to select comments that received more than 10 replies
query = """
SELECT
parent,
COUNT(id)
FROM
`bigquery-public-data.hacker_news.comments`
GROUP BY
parent
HAVING
COUNT(id) > 10
"""- We
GROUP BYtheparentcolumn andCOUNT()theidcolumn in order to figure out the number of comments that were made as responses to a specific comment. - Furthermore, since we're only interested in popular comments, we'll look at comments with more than 10 replies. So, we'll only return groups
HAVINGmore than 10 ID's.
The column resulting from COUNT(id) was called f0_. That's not a very descriptive name. We can change the name by adding AS NumPosts after the aggregation function. This is called aliasing.
If we are ever unsure what to put inside the COUNT() function, we can do COUNT(*) to count the rows in each group. It is more readable, because we know it's not focusing on other columns. It also scans less data than if supplied column names (making it faster and using less of our data access quota).
# improved version of earlier query, with aliasing & improved readability
query_improved = """
SELECT
parent,
COUNT(*) AS NumPosts
FROM
`bigquery-public-data.hacker_news.comments`
GROUP BY
parent
HAVING
COUNT(*) > 10
"""Order your results to focus on the most important data for your use case. #
Let's use the US Traffic Fatality Records database, which contains information on traffic accidents in the US where at least one person died. We'll investigate the accident_2015 table. We'll want to find which day of the week has the most fatal motor accidents.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# construct a reference to the "nhtsa_traffic_fatalities" dataset
dataset_ref = client.dataset("nhtsa_traffic_fatalities", project="bigquery-public-data")
# fetch the dataset (API request)
dataset = client.get_dataset(dataset_ref)# construct a reference to the "accident_2015" table
table_ref = dataset_ref.table("accident_2015")
# fetch the table (API request)
table = client.get_table(table_ref)- the
consecutive_numbercolumn contains a unique ID for each accident, and - the
timestamp_of_crashcolumn contains the date of the accident in DATETIME format,
To change the order of our results, like applying ordering to dates, we use some techniques.
ORDER BYis usually the last clause in the query, and it sorts the results returned by the rest of the query. We can reverse the order using theDESCargument (short for 'descending').- Dates come up very frequently in real-world databases. There are two ways that dates can be stored in BigQuery: as a
DATE, with the format of YYYY-MM-DD, or as aDATETIME, with the same format of date but with time added at the end.- Often we'll want to look at part of a date, like the year or the day. We do this with
EXTRACT. SQL is so clever. So, we can ask for information beyond just extracting part of the cell. For example,EXTRACT(WEEK from Date)returns the 1-53 week number for each YYYY-MM-DD in theDatecolumn. - More info: This link
- Often we'll want to look at part of a date, like the year or the day. We do this with
# query to find out the number of accidents for each day of the week
query = """
SELECT
COUNT(consecutive_number) AS num_accidents,
EXTRACT(DAYOFWEEK FROM timestamp_of_crash) AS day_of_week
FROM
`bigquery-public-data.nhtsa_traffic_fatalities.accident_2015`
GROUP BY
day_of_week
ORDER BY
num_accidents DESC
"""- We
EXTRACTthe day of the week (asday_of_week) from thetimestamp_of_crashcolumn, and GROUP BYthe day of the week, before weCOUNTtheconsecutive_numbercolumn to determine the number of accidents for each day of the week.- Notice that the data is sorted descending by the
num_accidentscolumn, where the days with more traffic accidents appear first. - Based on the BigQuery documentation, the
DAYOFWEEKfunction returns "an integer between 1 (Sunday) and 7 (Saturday), inclusively".
Organize your query for better readability. This becomes especially important for complex queries. #
We're going to find out how many Bitcoin transactions were made each day for the entire timespan of a bitcoin transaction dataset. We'll use Bitcoin Blockchain Historical Data and investigate the transactions table.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# construct a reference to the "crypto_bitcoin" dataset
dataset_ref = client.dataset("crypto_bitcoin", project="bigquery-public-data")
# fetch the dataset (API request)
dataset = client.get_dataset(dataset_ref)# construct a reference to the "transactions" table
table_ref = dataset_ref.table("transactions")
# fetch the table (API request)
table = client.get_table(table_ref)AS is a convenient way to clean up the data and rename the columns generated by our query, which is also known as aliasing. It's even more powerful when combined with WITH in what's called a "common table expression" (or CTE), which is a temporary table that it returns within our query. CTEs are helpful for splitting the queries into readable chunks, and writing queries against them. CTEs let us shift a lot of our data cleaning into SQL. That's an especially good thing in the case of BigQuery, because it is vastly faster than doing the work in Pandas.
It's important to note that CTEs only exist inside the query where we create them, and we can't reference them in later queries.
# query to select the number of transactions per date, sorted by date
query = """
WITH time AS (
SELECT
EXTRACT(DATE FROM block_timestamp) AS trans_date
FROM
`bigquery-public-data.crypto_bitcoin.transactions`
)
SELECT
COUNT(*) AS transactions,
trans_date
FROM
time
GROUP BY
trans_date
ORDER BY
trans_date
"""- Since the
block_timestampcolumn contains the date of each transaction in DATETIME format, we'll convert these into DATE format using theDATE()command. We do that using a CTE. - The next part of the query counts the number of transactions for each date and sorts the table so that earlier dates appear first.
- Since the output returned sorted, we can easily plot the raw results to show us the number of Bitcoin transactions per day over the whole timespan of this dataset.
# query that shows, for each hour of the day, the corresponding number of trips and average speed
query = """
WITH RelevantRides AS (
SELECT
EXTRACT(HOUR FROM trip_start_timestamp) AS hour_of_day,
trip_miles,
trip_seconds
FROM
`bigquery-public-data.chicago_taxi_trips.taxi_trips`
WHERE
EXTRACT(DATE FROM trip_start_timestamp) BETWEEN '2017-01-01' AND '2017-07-01'
AND trip_seconds > 0
AND trip_miles > 0
)
SELECT
hour_of_day,
COUNT(*) AS num_trips,
3600 * SUM(trip_miles) / SUM(trip_seconds) AS avg_mph
FROM
RelevantRides
GROUP BY
hour_of_day
ORDER BY
hour_of_day
"""Combine data sources. Critical for almost all real-world data problems. #
GitHub is the most popular place to collaborate on software projects. A GitHub repository is a collection of files associated with a specific project. Most repos on GitHub are shared under a specific legal license, which determines the legal restrictions on how they are used. We're going to find how many files are covered by each type of software license. We'll work with two tables in the database.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# construct a reference to the "github_repos" dataset
dataset_ref = client.dataset("github_repos", project="bigquery-public-data")
# fetch the dataset (API request)
dataset = client.get_dataset(dataset_ref)# construct a reference to the "licenses" table
licenses_ref = dataset_ref.table("licenses")
# fetch the table (API request)
licenses_table = client.get_table(licenses_ref)- The first table is the
licensestable, which provides the name of each GitHub repo (in therepo_namecolumn) and its corresponding license.
# construct a reference to the "sample_files" table
files_ref = dataset_ref.table("sample_files")
# fetch the table (API request)
files_table = client.get_table(files_ref)- The second table is the
sample_filestable, which provides the GitHub repo that each file belongs to (in therepo_namecolumn).
We have the tools to obtain data from a single table in whatever format we want it. But what if the data we want is spread across multiple tables? That's where JOIN comes in! We combine information from both tables by matching rows where a column in one table matches a column in another table.
# query to determine the number of files per license, sorted by number of files
query = """
SELECT
L.license,
COUNT(*) AS number_of_files
FROM
`bigquery-public-data.github_repos.sample_files` AS sf
INNER JOIN `bigquery-public-data.github_repos.licenses` AS L ON sf.repo_name = L.repo_name
GROUP BY
L.license
ORDER BY
number_of_files DESC
"""ONdetermines which column in each table to use to combine the tables.- There are some types of
JOIN, but anINNER JOINis very widely used, which means that a row will only be put in the final output table if the value in the columns we're using to combine them shows up in both the tables we're joining.
A lot of the data are text. There is a great technique we can apply to these texts. A WHERE clause can limits out results to rows with certain text using the LIKE feature. To select rows with 'bigquery' in tag column:
query = """
SELECT
id,
title,
owner_user_id
FROM
`bigquery-public-data.stackoverflow.posts_questions`
WHERE
tags LIKE '%bigquery%'
"""%is a wildcard and is used to include rows where there is other text in addition to the word "bigquery" (e.g., if a row has atag"bigquery-sql").
We can generalize the query application by defining a python function and add the parameters in {} in our query.
# a general function to get experts on any topic
def expert_finder(topic, client):
"""
Returns a DataFrame with the user IDs who have written Stack Overflow answers on a topic.
"""
query = """
SELECT
a.owner_user_id AS user_id,
COUNT(*) AS number_of_answers
FROM
`bigquery-public-data.stackoverflow.posts_questions` AS q
INNER JOIN `bigquery-public-data.stackoverflow.posts_answers` AS a ON q.id = a.parent_Id
WHERE
q.tags LIKE '%{topic}%'
GROUP BY
a.owner_user_id
"""
safe_config = bigquery.QueryJobConfig(maximum_bytes_billed=10 ** 9)
my_query_job = client.query(query, job_config=safe_config)
results = my_query_job.to_dataframe()
return resultsPractice with Kaggle BigQuery datasets available here.
Take your SQL skills to the next level.
Combine information from multiple tables. #
We'll work with the Hacker News dataset. We want to pull information from the stories and comments tables.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# construct a reference to the "hacker_news" dataset
dataset_ref = client.dataset("hacker_news", project="bigquery-public-data")
# fetch the dataset (API request)
dataset = client.get_dataset(dataset_ref)- If you are familiar with the data, it's not neccessary to construct references to the tables and fetch them.
Recall that we can use an INNER JOIN to pull rows from both tables where the value in a column of the left table has a match in a column of the right table ("left" to the table that appears before the JOIN in the query). A LEFT JOIN returns all rows where the two tables have matching entries, along with all of the rows in the left table (whether there is a match or not). If we instead use a RIGHT JOIN, we get the matching rows, along with all rows in the right table (whether there is a match or not). Finally, a FULL JOIN returns all rows from both tables. Note that in general, any row that does not have a match in both tables will have NULL entries for the missing values.
As you've seen, JOINs horizontally combine results from different tables. If we instead would like to vertically concatenate columns, we can do so with a UNION. Note that with a UNION, the data types of both columns must be the same. We use UNION ALL to include duplicate values in the concatenated results. If we'd like to drop duplicate values, we use UNION DISTINCT in the query.
We want to create a table showing all stories posted on January 1, 2012, along with the corresponding number of comments. We use a LEFT JOIN so that the results include stories that didn't receive any comments.
# query to select all stories posted on January 1, 2012, with number of comments
query = """
WITH c AS (
SELECT
parent,
COUNT(*) AS num_comments
FROM
`bigquery-public-data.hacker_news.comments`
GROUP BY
parent
)
SELECT
s.id AS story_id,
s.by,
s.title,
c.num_comments
FROM
`bigquery-public-data.hacker_news.stories` AS s
LEFT JOIN c ON s.id = c.parent
WHERE
EXTRACT(DATE FROM s.time_ts) = '2012-01-01'
ORDER BY
c.num_comments DESC
"""
# run the query (if it's less than 1 GB), and return a pandas DataFrame
safe_config = bigquery.QueryJobConfig(maximum_bytes_billed=10 ** 9)
join_result = client.query(query, job_config=safe_config).result().to_dataframe()- Since the results are ordered by the
num_commentscolumn, stories without comments appear at the end of the DataFrame. (Remember thatNaNstands for "not a number".)
Next, we write a query to select all usernames corresponding to users who wrote stories or comments on January 1, 2014. We use UNION DISTINCT (instead of UNION ALL) to ensure that each user appears in the table at most once.
# query to select all users who posted stories or comments on January 1, 2014
query = """
SELECT
c.by
FROM
`bigquery-public-data.hacker_news.comments` AS c
WHERE
EXTRACT(DATE FROM c.time_ts) = '2014-01-01'
UNION
DISTINCT
SELECT
s.by
FROM
`bigquery-public-data.hacker_news.stories` AS s
WHERE
EXTRACT(DATE FROM s.time_ts) = '2014-01-01'
"""
# run the query (if it's less than 1 GB), and return a pandas DataFrame
safe_config = bigquery.QueryJobConfig(maximum_bytes_billed=10 ** 9)
union_result = client.query(query, job_config=safe_config).result().to_dataframe()- To get the number of users who posted on January 1, 2014, we need only take the length of the DataFrame.
Perform complex calculations on groups of rows. #
Analytic functions, same as aggregate functions, operate on a set of rows. However, unlike aggregate functions, analytic functions return a (potentially different) value for each row in the original table.
All analytic functions have an OVER clause, which defines the sets of rows used in each calculation. The OVER clause has three (optional) parts:
- The
PARTITION BYclause divides the rows of the table into different groups. - The
ORDER BYclause defines an ordering within each partition. - The window frame clause identifies the set of rows used in each calculation (Analytic functions are sometimes referred to as window functions!).
There are many ways to write window frame clauses. Some examples are:
ROWS BETWEEN 1 PRECEDING AND CURRENT ROW- the previous row and the current row.ROWS BETWEEN 3 PRECEDING AND 1 FOLLOWING- the 3 previous rows, the current row, and the following row.ROWS BETWEEN UNBOUNDED PRECEDING AND UNBOUNDED FOLLOWING- all rows in the partition.
BigQuery supports a wide variety of analytic functions, and we'll explore a few here.
- Aggregate Functions take all of the values within the window as input and return a single value. The
OVERclause is what ensures that it's treated as an analytic (aggregate) function.MIN()orMAX()AVG()orSUM()COUNT()
- Navigation Functions assign a value based on the value in a (usually) different row than the current row.
LEAD()(andLAG()) returns the value on a subsequent (or preceding) row.FIRST_VALUE()(orLAST_VALUE()) returns the first (or last) value in the input.
- Numbering Functions assign integer values to each row based on the ordering.
ROW_NUMBER()returns the order in which rows appear in the input (starting with 1).RANK()- All rows with the same value in the ordering column receive the same rank value, where the next row receives a rank value which increments by the number of rows with the previous rank value.
We'll work with the San Francisco Open Data dataset. We want explore data from the bikeshare_trips table.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# construct a reference to the "san_francisco" dataset
dataset_ref = client.dataset("san_francisco", project="bigquery-public-data")
# fetch the dataset (API request)
dataset = client.get_dataset(dataset_ref)Each row of the table corresponds to a different bike trip, and we can use an analytic function to calculate the cumulative number of trips for each date in 2015.
# query to count the (cumulative) number of trips per day
num_trips_query = """
WITH trips_by_day AS (
SELECT
DATE(start_date) AS trip_date,
COUNT(*) AS num_trips
FROM
`bigquery-public-data.san_francisco.bikeshare_trips`
WHERE
EXTRACT(YEAR FROM start_date) = 2015
GROUP BY
trip_date
)
SELECT
*,
SUM(num_trips) OVER (
ORDER BY
trip_date ROWS BETWEEN UNBOUNDED PRECEDING AND CURRENT ROW
) AS cumulative_trips
FROM
trips_by_day
"""- The query uses a common table expression (CTE) to first calculate the daily number of trips. Then, we use
SUM()as an aggregate function. - Since there is no
PARTITION BYclause, the entire table is treated as a single partition. - The
ORDER BYclause orders the rows by date, where earlier dates appear first. - By setting the window frame clause to
ROWS BETWEEN UNBOUNDED PRECEDING AND CURRENT ROW, we ensure that all rows up to and including the current date are used to calculate the (cumulative) sum. (BTW, This is the default behavior. So, we can skip this part.)
We can track the stations where each bike began (in start_station_id) and ended (in end_station_id) the day on October 25, 2015.
# query to track beginning and ending stations on October 25, 2015, for each bike
start_end_query = """
SELECT
bike_number,
TIME(start_date) AS trip_time,
FIRST_VALUE(start_station_id) OVER (
PARTITION BY bike_number
ORDER BY
start_date ROWS BETWEEN UNBOUNDED PRECEDING AND UNBOUNDED FOLLOWING
) AS first_station_id,
LAST_VALUE(end_station_id) OVER (
PARTITION BY bike_number
ORDER BY
start_date ROWS BETWEEN UNBOUNDED PRECEDING AND UNBOUNDED FOLLOWING
) AS last_station_id,
start_station_id,
end_station_id
FROM
`bigquery-public-data.san_francisco.bikeshare_trips`
WHERE
DATE(start_date) = '2015-10-25'
"""- The query uses both
FIRST_VALUE()andLAST_VALUE()as analytic functions. - The
PARTITION BYclause breaks the data into partitions based on thebike_numbercolumn. Since this column holds unique identifiers for the bikes, this ensures the calculations are performed separately for each bike. - The
ORDER BYclause puts the rows within each partition in chronological order. - Since the window frame clause is
ROWS BETWEEN UNBOUNDED PRECEDING AND UNBOUNDED FOLLOWING, for each row, its entire partition is used to perform the calculation. (This ensures the calculated values for rows in the same partition are identical.)
We want to know about the length of the break (in minutes) that a driver had before each trip started (this corresponds to the time between trip_start_timestamp of the current trip and trip_end_timestamp of the previous trip).
# query to track break time on May 1, 2017, for each taxi driver
break_time_query = """
SELECT
taxi_id,
trip_start_timestamp,
trip_end_timestamp,
TIMESTAMP_DIFF(
trip_start_timestamp,
LAG(trip_end_timestamp) OVER (
PARTITION BY
taxi_id
ORDER BY
trip_start_timestamp
),
MINUTE
) AS prev_break
FROM
`bigquery-public-data.chicago_taxi_trips.taxi_trips`
WHERE
DATE(trip_start_timestamp) = '2017-05-01'
"""- The
TIMESTAMP_DIFF()function takes three arguments. This function provides the time difference of the timestamps in the first two arguments. We should use theLAG()function to pull the timestamp corresponding to the end of the previous trip (for the sametaxi_id).
Learn to query complex datatypes in BigQuery. #
We can organize our information in many different tables. Another option in BigQuery is to organize all of the information in a single table. In this case, all of the information from related tables is collapsed into a single column in main table. We refer to that column as a nested column. Nested columns have type RECORD (or STRUCT) in the table schema. Each entry in a nested field is like a dictionary. To query a column with nested data, we need to use . notation to identify each field in that column.
Now consider the case where for example a pet can have multiple toys, or multiple owners. In this case, to collapse this information into a single column, we need a column contains REPEATED (or ARRAY) data, because it permits more than one value for each row. Each entry in a repeated field is like an ordered list of values with the same datatype. When querying repeated data, we need to put the name of the column containing the repeated data inside an UNNEST() function. This flattens the repeated data so that we have one element on each row.
But in the real world examples, we have nested columns with repeated data.
We'll work with the Google Analytics Sample dataset. It contains information tracking the behavior of visitors to the Google Merchandise store. We work with ga_sessions_20170801 table, which has many nested fields. For a description of each field, refer to this data dictionary.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# construct a reference to the "google_analytics_sample" dataset
dataset_ref = client.dataset("google_analytics_sample", project="bigquery-public-data")# construct a reference to the "ga_sessions_20170801" table
table_ref = dataset_ref.table("ga_sessions_20170801")
# fetch the table (API request)
table = client.get_table(table_ref)# print schema field for the 'device' column
print(table.schema[7])We refer to the browser field (which is nested in the device column) and the transactions field (which is nested inside the totals column) to count the number of transactions per browser.
# query to count the number of transactions per browser
query = """
SELECT
device.browser AS device_browser,
SUM(totals.transactions) AS total_transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_20170801`
GROUP BY
device_browser
ORDER BY
total_transactions DESC
"""- By storing the information in the
deviceandtotalscolumns asRECORDs (as opposed to separate tables), we avoid expensiveJOINs. This increases performance and keeps us from having to worry aboutJOINkeys (and which tables have the exact data we need).
Now we'll work with the hits column as an example of data that is both nested and repeated. Since:
hitsis aRECORD(contains nested data) and isREPEATED,hitNumber,page, andtypeare all nested inside thehitscolumn, andpagePathis nested inside thepagefield,
# query to determine most popular landing point on the website
query = """
SELECT
hits.page.pagePath AS path,
COUNT(hits.page.pagePath) AS counts
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_20170801`,
UNNEST(hits) AS hits
WHERE
hits.type = "PAGE"
AND hits.hitNumber = 1
GROUP BY
path
ORDER BY
counts DESC
"""Write queries to run faster and use less data. #
Sometimes it doesn't matter whether your query is efficient or not. For example, a query you expect to run only once, or it might be working on a small dataset. But what about queries that will be run many times, like a query that feeds data to a website? Or what about queries on huge datasets?
We will use two functions to compare the efficiency of different queries.
# create a "Client" object
from google.cloud import bigquery
client = bigquery.Client()# function shows the amount of data the query uses
def show_amount_of_data_scanned(query):
# dry_run lets us see how much data the query uses without running it
dry_run_config = bigquery.QueryJobConfig(dry_run=True)
query_job = client.query(query, job_config=dry_run_config)
print(
"Data processed: {} GB".format(
round(query_job.total_bytes_processed / 10 ** 9, 3)
)
)# function shows how long it takes for the query to execute
from time import time
def show_time_to_run(query):
time_config = bigquery.QueryJobConfig(use_query_cache=False)
start = time()
query_result = client.query(query, job_config=time_config).result()
end = time()
print("Time to run: {} seconds".format(round(end - start, 3)))It is tempting to start queries with SELECT * FROM. This is especially important if there are text fields that you don't need, because text fields tend to be larger than other fields.
star_query = "SELECT * FROM `bigquery-public-data.github_repos.contents`"
show_amount_of_data_scanned(star_query)
>>> Data processed: 2471.537 GBbasic_query = "SELECT size, binary FROM `bigquery-public-data.github_repos.contents`"
show_amount_of_data_scanned(basic_query)
Data processed: 2.371 GBmore_data_query = """
SELECT
MIN(start_station_id) AS start_station_name,
MIN(end_station_id) AS end_station_name,
AVG(duration_sec) AS avg_duration_sec
FROM
`bigquery-public-data.san_francisco.bikeshare_trips`
WHERE
start_station_id != end_station_id
GROUP BY
start_station_id,
end_station_id
LIMIT
10
"""
show_amount_of_data_scanned(more_data_query)
>>> Data processed: 0.076 GBless_data_query = """
SELECT
start_station_name,
end_station_name,
AVG(duration_sec) AS avg_duration_sec
FROM
`bigquery-public-data.san_francisco.bikeshare_trips`
WHERE
start_station_name != end_station_name
GROUP BY
start_station_name,
end_station_name
LIMIT
10
"""
show_amount_of_data_scanned(less_data_query)
>>> Data processed: 0.06 GB- Since there is a 1:1 relationship between the station ID and the station name, we don't need to use the
start_station_idandend_station_idcolumns in the query. By using only the columns with the station names, we scan less data.
In 1:1 JOINs, each row in each table has at most one match in the other table. In N:1 JOIN, each row in one table matches potentially many rows in the other table. Finally, in N:N JOIN, a group of rows in one table can match a group of rows in the other table.
big_join_query = """
SELECT
repo,
COUNT(DISTINCT c.committer.name) AS num_committers,
COUNT(DISTINCT f.id) AS num_files
FROM
`bigquery-public-data.github_repos.commits` AS c,
UNNEST(c.repo_name) AS repo
INNER JOIN `bigquery-public-data.github_repos.files` AS f ON f.repo_name = repo
WHERE
f.repo_name IN ('tensorflow/tensorflow', 'twbs/bootstrap', 'Microsoft/vscode', 'torvalds/linux')
GROUP BY
repo
ORDER BY
repo
"""
show_time_to_run(big_join_query)
>>> Time to run: 7.192 secondssmall_join_query = """
WITH commits AS (
SELECT
COUNT(DISTINCT committer.name) AS num_committers,
repo
FROM
`bigquery-public-data.github_repos.commits`,
UNNEST(repo_name) AS repo
WHERE
repo IN ('tensorflow/tensorflow', 'twbs/bootstrap', 'Microsoft/vscode', 'torvalds/linux')
GROUP BY
repo
),
files AS (
SELECT
COUNT(DISTINCT id) AS num_files,
repo_name AS repo
FROM
`bigquery-public-data.github_repos.files`
WHERE
repo_name IN ('tensorflow/tensorflow', 'twbs/bootstrap', 'Microsoft/vscode', 'torvalds/linux')
GROUP BY
repo
)
SELECT
commits.repo,
commits.num_committers,
files.num_files
FROM
commits
INNER JOIN files ON commits.repo = files.repo
ORDER BY
repo
"""
show_time_to_run(small_join_query)
>>> Time to run: 5.245 secondsExtract human-understandable insights from any machine learning model.
Why and when do you need insights? #
- What features in the data did the model think are most important?
- For any single prediction from a model, how did each feature in the data affect that particular prediction?
- How does each feature affect the model's predictions in a big-picture sense (what is its typical effect when considered over a large number of possible predictions)?
- Debugging - Dirty data, preprocessing codes, and data leakage are potential sources for bugs. Debugging is one of the most valuable skills in data science. Understanding the patterns a model is finding will help us identify when those are at odds with our knowledge of the real world, and this is typically the first step in tracking down bugs.
- Informing Feature Engineering - Feature engineering is usually the most effective way to improve model accuracy. The techniques we'll learn would make it transparent what features are more important. As an increasing number of datasets start with 100s or 1000s of raw features, this approach is becoming increasingly important.
- Directing Future Data Collection - Collecting new types of data can be expensive, so businesses and organizations only want to do this if they know it will be worthwhile. Model-based insights give us a good understanding of the value of features we currently have, which will help us reason about what new values may be most helpful.
- Informing Human Decision-making - Some decisions are made automatically by models. But many important decisions are made by humans. For these decisions, insights can be more valuable than predictions.
- Building Trust - Many people won't assume they can trust our model for important decisions without verifying some basic facts. Showing insights that fit their general understanding of the problem will help build trust, even among people with little deep knowledge of data science.
What features does your model think are important? #
What features have the biggest impact on predictions? This concept is called feature importance. There are multiple ways to measure feature importance. We'll focus on permutation importance (PI). Compared to most other approaches, PI is:
- fast to calculate,
- widely used and understood, and
- consistent with properties we would want a feature importance measure to have.
Our data includes useful features, features with little predictive power, as well as some other features we won't focus on. PI is calculated after a model has been fitted. The key question is: If I randomly shuffle a single column of the validation data, leaving the target and all other columns in place, how would that affect the accuracy of predictions in that now-shuffled data? Model accuracy especially suffers if we shuffle a column that the model relied on heavily for predictions.
We will use a model that predicts whether a football team will have the Man of the Game winner based on the team's statistics. Model-building isn't our current focus, so the cell below loads the data and builds a rudimentary model.
# load and prepare data
import pandas as pd
data = pd.read_csv("../input/fifa-2018-match-statistics/FIFA 2018 Statistics.csv")
# convert target from string "Yes"/"No" to binary
y = data["Man of the Match"] == "Yes"
import numpy as np
base_features = [i for i in data.columns if data[i].dtype in [np.int64]]
X = data[base_features]# split train and valid data
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(X, y, random_state=1)# define & fit model
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=100, random_state=0).fit(X_train, y_train)We'll use elib library to calculate and show importances.
# calculate importances
from eli5.sklearn import PermutationImportance
perm = PermutationImportance(model, random_state=1).fit(X_valid, y_valid)
# show them
from eli5 import show_weights
show_weights(perm, feature_names=X_valid.columns.tolist())| Weight | Feature |
|---|---|
| 0.1750 ± 0.0848 | Goal Scored |
| 0.0500 ± 0.0637 | Distance Covered (Kms) |
| 0.0437 ± 0.0637 | Yellow Card |
| 0.0187 ± 0.0637 | Free Kicks |
| 0.0187 ± 0.0637 | Fouls Committed |
| 0.0125 ± 0.0637 | Pass Accuracy % |
| 0.0063 ± 0.0612 | Saves |
| -0.0187 ± 0.0306 | Attempts |
| -0.0500 ± 0.0637 | Passes |
The values towards the top are the most important features, and those towards the bottom matter least. The number in each row shows how much model performance decreased with a random shuffling (in this case, using "accuracy" as the performance metric). The amount of randomness in our PI is calculated by repeating the process with multiple shuffles and the number after the ± shows that.
We'll occasionally see negative values for PIs. This happens when the feature didn't matter, but random chance caused the predictions on shuffled data to be more accurate. This is more common with small datasets, because there is more room for luck/chance.
- Higher PI for a feature can have multiple reasons, not just one. It's not a direct path!
- We can create better features, using
abs_lat_changeinstead of onlypickup_latitudeanddropoff_latitude. - The scale of features does not affect PI. The only reason that rescaling a feature would affect PI is indirectly, if rescaling helped or hurt the ability of the particular learning method we're using to make use of that feature. That won't happen with tree based models, like the Random Forest.
How does each feature affect your predictions? #
While feature importance shows what variables most affect predictions, partial dependence plots show how a feature affects predictions. This is useful to answer questions like:
- Controlling for all other house features, what impact do longitude and latitude have on home prices? To restate this, how would similarly sized houses be priced in different areas?
- Are predicted health differences between two groups due to differences in their diets, or due to some other factor?
Partial dependence plots can be interpreted similarly to the coefficients in linear or logistic regression models.
Like permutation importance, partial dependence plots are calculated after a model has been fit.
We repeatedly alter the value for one variable and use the fitted model to make a series of predictions. We trace out predicted outcomes as we move from small values to large ones. Interactions between features may cause the plot for the dataset.
We can use decision tree.
# define and fit model
from sklearn.tree import DecisionTreeClassifier
tree_model = DecisionTreeClassifier(
random_state=0, max_depth=5, min_samples_split=5
).fit(X_train, y_train)# export decision tree
from sklearn.tree import export_graphviz
tree_graph = export_graphviz(tree_model, out_file=None, feature_names=base_features)
# show it
import graphviz
graphviz.Source(tree_graph)- Leaves with children show their splitting criterion on the top.
- The pair of values at the bottom show the count of False values and True values for the target respectively, of data points in that node of the tree.
Or, we can use PDPBox library to create a partial dependence plot.
# define and fit model
from sklearn.ensemble import RandomForestClassifier
rf_model = RandomForestClassifier(random_state=0).fit(train_X, train_y)feature_to_plot = "Distance Covered (Kms)"
# create data
from pdpbox import pdp, get_dataset, info_plots
pdp_iso = pdp.pdp_isolate(
model=rf_model,
dataset=X_valid,
model_features=base_features,
feature=feature_to_plot,
)
# plot it
pdp.pdp_plot(pdp_iso, feature_to_plot)
from matplotlib import pyplot as plt
plt.show()- The y axis is interpreted as change in the prediction from what it would be predicted at the baseline or leftmost value.
- A shaded area indicates the level of confidence.
Or we can use 2D partial dependence plots to clarify about interactions between features.
features_to_plot = ["Goal Scored", "Distance Covered (Kms)"]
# create data
pdp_inter = pdp.pdp_interact(
model=rf_model,
dataset=X_valid,
model_features=base_features,
features=features_to_plot,
)
# plot it
pdp.pdp_interact_plot(
pdp_interact_out=pdp_inter, feature_names=features_to_plot, plot_type="contour"
)
plt.show()- It is similar to previous PDP plot except we use
pdp_interactinstead ofpdp_isolateandpdp_interact_plotinstead ofpdp_isolate_plot.
Consider a scenario where we have only 2 predictive features, feat_A and feat_B. Both have minimum values of -1 and maximum values of 1. The partial dependence plot for feat_A increases steeply over its whole range, whereas the partial dependence plot for feat_B increases at a slower rate (less steeply) over its whole range. Does this guarantee that feat_A will have a higher permutation importance than feat_B?
- No. For example,
feat_Acould have a big effect in the cases where it varies, but could have a single value 99% of the time. In that case, permutingfeat_Awouldn't matter much, since most values would be unchanged.
Understand individual predictions #
We've used techniques to extract general insights from a machine learning model. SHAP Values (an acronym from SHapley Additive exPlanations) break down a prediction to show the impact of each feature. Where could we use this?
- A model says a bank shouldn't loan someone money, and the bank is legally required to explain the basis for each loan rejection.
- A healthcare provider wants to identify what factors are driving each patient's risk of some disease so they can directly address those risk factors with targeted health interventions.
SHAP values interpret/decompose the impact of having certain values for given features in comparison to the prediction we'd make if those features took some baseline value.
There is some complexity to the technique, to ensure that the baseline plus the sum of individual effects adds up to the prediction. This blog post has a theoretical explanation about it.
# define & fit model
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(random_state=0).fit(X_train, y_train)# look at the raw predictions for a single row, we can do for multiple
data_for_prediction = X_valid.iloc[0, :]
# convert data column to a row, -1 is for adaptivity
data_for_prediction_array = data_for_prediction.values.reshape(1, -1)
# find predictions' probabilities
my_model.predict_proba(data_for_prediction_array)
>>> array([[0.3, 0.7]])- The team is 70% likely to have a player win the award.
We calculate SHAP values using the SHAP library.
# create object can calculate SHAP values
import shap
explainer = shap.TreeExplainer(model)
# calculate SHAP values
shap_values = explainer.shap_values(data_for_prediction)- The
shap_valuesobject is a list with two arrays. The first array is the SHAP values for a negative outcome (don't win the award), and the second array is the list of SHAP values for the positive outcome (wins the award). - We typically think about predictions in terms of the prediction of a positive outcome, so we'll pull out
shap_values[1].
# visualize
shap.initjs()
shap.force_plot(explainer.expected_value[1], shap_values[1], data_for_prediction)- For
output valuewe predicted 0.7, whereas thebase valueis 0.4979. - The biggest impact comes from
Goal Scoredbeing 2. Though theBall Possessionvalue has a meaningful effect decreasing the prediction. - If you subtract the length of the blue bars from the length of the pink bars, it equals the distance from the base value to the output.
The SHAP package has explainers for every type of model.
shap.TreeExplainerworks with tree models.shap.DeepExplainerworks with deep learning models.shap.KernelExplainerworks with all models, though it is slower than other explainers and it offers an approximation rather than exact SHAP values.
It seems time_in_hospital doesn't matter in readmission prediction. If that is what our model concluded, the doctors will believe it. But could the data be wrong, or is our model doing something more complex than they expect? If we'd like to show the raw readmission rate for each value of time_in_hospital to see how it compares to the partial dependence plot:
all_train = pd.concat([X_train, y_train], axis=1)
all_train.groupby("time_in_hospital")["readmitted"].mean().plot()Aggregate SHAP values for even more detailed model insights #
We'll expand on SHAP values, seeing how aggregating many SHAP values can give more detailed alternatives to permutation importance and partial dependence plots. We will focus on two of SHAP libarary's visualizations. These visualizations have conceptual similarities to permutation importance and partial dependence plots.
# define & fit model
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(random_state=0).fit(X_train, y_train)Permutation importance is great because it created simple numeric measures to see which features mattered to a model. But it doesn't tell us how each features matter. If a feature has medium permutation importance, that could mean it has
- a large effect for a few predictions, but no effect in general, or
- a medium effect for all predictions.
SHAP Summary Plots give us a birds-eye view of feature importance and what is driving it.
# create object can calculate SHAP values
import shap
explainer = shap.TreeExplainer(model)
# calculate SHAP values for all of X_valid rather than a single row
shap_values = explainer.shap_values(X_valid)# visualize
shap.summary_plot(shap_values[1], X_valid)- For classification problems, there is a separate array of SHAP values for each possible outcome.
shap_values[0]for the prediction ofFalseandshap_values[1]forTrue. - Calculating SHAP values can be slow especially with reasonably sized datasets. The exception is when using an xgboost model, which SHAP has some optimizations for and which is thus much faster.
- Vertical location shows what feature it is depicting.
- Color shows whether that feature was high or low for that row of the dataset.
- Horizontal location shows whether the effect of that value caused a higher or lower prediction.
- Because the range of effects (distance between smallest effect and largest effect) is so sensitive to outliers, permutation importance is a better measure of what's generally important to the model. However if all dots on the graph are widely spread from each other, that is a reasonable indication that permutation importance is high.
- The jumbling (have blue and pink dots jumbled together) suggests that the variable has an interaction effect with other variables. We could investigate that with SHAP Contribution Dependence Plots.
Some things we should be able to easily pick out:
- High values of
Goal Scoredcaused higher predictions, and low values caused low predictions
We've previously used Partial Dependence Plots to show how a single feature impacts predictions. But what is the distribution of effects? SHAP Dependence Contribution Plots provide a similar insight to PDP's, but they add a lot more detail.
# visualize
shap.dependence_plot(
"Ball Possession %", shap_values[1], X_valid, interaction_index="Goal Scored"
)- There is only line that's different from the
summary_plot. - If we don't supply an argument for
interaction_index, SHAP library uses some logic to pick one that may be interesting.
- Each dot represents a row of the data. The horizontal location is the actual value from the dataset, and the vertical location shows what having that value did to the prediction.
- The fact this slopes upward says that the more we possess the ball, the higher the model's prediction is for winning the "Man of the Game" award.
- The spread suggests that other features must interact with
Ball Possession %. For example, points with same ball possession value and different SHAP values. - This suggests we delve into the interactions, and the plots include color coding to help do that.
- Consider those two points from bottom right stand out spatially as being far away from the upward trend. They are both indicating the team scored one goal. We can interpret this to say In general, having the ball increases a team's chance of having their player win the award. But if they only score one goal, that trend reverses and the award judges may penalize them for having the ball so much if they score that little.
- The SHAP vaue is an estimate of the impact of a given feature on the prediction. So, if the dots trend from upper left to lower right, that means low values of
feature_of_interestcause higher predictions. So increasingfeature_of_interesthas a more positive impact on predictions whenother_featureis high.
Distinguish yourself by learning to work with text data.
Get started with NLP. #
We will use spaCy, the leading Natural Language Processing (NLP) library, to take on some of the most important tasks in working with text.
- Basic text processing and pattern matching.
- Building machine learning models with text.
- Representing text with word embeddings that numerically capture the meaning of words and documents.
spaCy relies on models that are language-specific. We can load a spaCy model with spacy.load or spacy.blank.
# load the English language model
import spacy
nlp = spacy.load("en")With the model loaded, we can process texts.
# tokenize text
doc = nlp("Tea is healthy and calming, don't you think?")- This returns a document object that contains tokens. A token is a unit of text in the document, such as individual words and punctuation. SpaCy splits contractions like "don't" into two tokens, "do" and "n't".
tokens = [token for token in doc]- Iterating through a document gives us token objects. Each of these tokens comes with additional information, such as
token.lemma_andtoken.is_stop. - The lemma of a word is its base form. For example, "walk" is the lemma of the word "walking".
token.lemma_returns the lemma. - Stopwords are words that occur frequently in the language and don't contain much information. English stopwords include "the", "is", "and", "but", "not".
token.is_stopreturns a booleanTrueif the token is a stopword (andFalseotherwise).
Language data has a lot of noise mixed in with informative content. Removing stop words might help the predictive model focus on relevant words. Lemmatizing helps by combining multiple forms of the same word into one base form ("calming", "calms", "calmed" would all change to "calm"). However, lemmatizing and dropping stopwords might result in our models performing worse. So we should treat this preprocessing as part of our hyperparameter optimization process.
Another common NLP task is matching tokens or phrases within chunks of text or whole documents. To match individual tokens, we create a Matcher and for a list of terms, it's easier and more efficient to use PhraseMatcher.
# create a PhraseMatcher object
from spacy.matcher import PhraseMatcher
matcher = PhraseMatcher(nlp.vocab, attr="LOWER")nlp.vocabis the tokenizer, using the vocabulary of our model.- Setting
attr="LOWER"will match the phrases on lowercased text and provides case insensitive matching.
The PhraseMatcher needs the patterns as document objects.
# create patterns
terms = ["Galaxy Note", "iPhone 11", "iPhone XS", "Google Pixel"]
patterns = [nlp(item) for item in terms]# add rules to the PhraseMatcher object
matcher.add("TerminologyList", None, *patterns)- In the first argument we provide a name for the set of rules we're matching to.
- In the second argument we define special actions to take on matched words.
# tokenize text
doc = nlp(
"Glowing review overall, and some really interesting side-by-side "
"photography tests pitting the iPhone 11 Pro against the "
"Galaxy Note 10 Plus and last year’s iPhone XS and Google Pixel 3."
)
# # apply PhraseMatcher to tokenized text
matches = matcher(doc)matches
>>> [(3766102292120407359, 17, 19),
(3766102292120407359, 22, 24),
(3766102292120407359, 30, 32),
(3766102292120407359, 33, 35)]- The matches are tuples of the match id (
match[0]) and the positions of the start (match[1]) and end (match[2]) of the phrase.
# load data
import pandas as pd
data = pd.read_json("../input/nlp-course/restaurant.json")# add rules to the PhraseMatcher object
patterns = [nlp(item) for item in menu]
matcher.add("MENU", None, *patterns)# create item_ratings as a dictionary of lists of stars for food items in the menu
from collections import defaultdict
item_ratings = defaultdict(list)
for idx, review in data.iterrows():
doc = nlp(review["text"])
matches = matcher(doc)
found_items = set([doc[match[1] : match[2]] for match in matches])
for item in found_items:
item_ratings[str(item).lower()].append(review["stars"])# calculate the mean ratings for each menu item
from statistics import mean
mean_ratings = {item: mean(ratings) for item, ratings in item_ratings.items()}# calculate the number of reviews for each item
counts = {item: len(ratings) for item, ratings in item_ratings.items()}
# find the worst items + errors of calculation based on item counts
sorted_ratings = sorted(mean_ratings, key=mean_ratings.get)
for item in sorted_ratings[:10]:
print(
f"{item:20} Ave rating: {mean_ratings[item]:.2f} ± {1/(counts[item]**0.5):.2f} \tcount: {counts[item]}"
)- The less data we have for any specific item, the less we can trust that the average rating is the "real" sentiment of the customers. This is fairly common sense. If more people tell us the same thing, we're more likely to believe it. It's also mathematically sound. As the number of data points increases, the error on the mean decreases as
1/sqrt(n).
Combine machine learning with your newfound NLP skills. #
A common task in NLP is text classification. Examples include spam detection, sentiment analysis, and tagging customer queries.
We'll create a classifier that will detect spam messages.
# load and shuffle data, sampling with frac<1, upsampling with frac>1
import pandas as pd
spam = pd.read_csv("../input/nlp-course/spam.csv").sample(frac=1, random_state=1)Machine learning models don't learn from raw text data. Instead, we need to convert the text to something numeric. The simplest common representation is a variation of one-hot encoding. We represent each document as a vector of term frequencies for each term in the vocabulary. The vocabulary is built from all the tokens (terms) in the corpus (the collection of documents). This is called the BOW (Bag-of-Words) representation.
Another common representation is TF-IDF (Term Frequency - Inverse Document Frequency). TF-IDF is similar to bag of words except that each term count is scaled by the term's frequency in the corpus. Using TF-IDF can potentially improve our models.
Once we have our documents in a bag of words representation (or TF-IDF), we can use those vectors as input to any machine learning model. spaCy handles the bag of words conversion and building a simple linear model for us with the TextCategorizer class.
The TextCategorizer is a spaCy pipe. Pipes are classes for processing and transforming tokens. When we load a spaCy model with spacy.load, there are default pipes that perform part of speech tagging, entity recognition, and other transformations. When we run text through a model, the output of the pipes are attached to the tokens in the doc object. When we create a blank spaCy model with spacy.blank, there is only a tokenizer pipe. We can remove or add pipes to models.
# create an empty model
import spacy
nlp = spacy.blank("en")# create the TextCategorizer
textcat = nlp.create_pipe(
"textcat", config={"exclusive_classes": True, "architecture": "bow"}
)
# add the TextCategorizer to the empty model
nlp.add_pipe(textcat)"exclusive_classes"makes categories mutually exclusive."architecture"configures model architecture:"ensemble"- Stacked ensemble of a bag-of-words model and a CNN model."simple_cnn"- A neural network model where token vectors are calculated using a CNN. The vectors are mean pooled and used as features in a feed-forward network."bow"- An ngram bag-of-words model. The features extracted can be controlled using the keyword argumentsngram_sizeandattr(ex."lower"). This architecture should run much faster than the others, but may not be as accurate, especially if texts are short.
# add labels to TextCategorizer
textcat.add_label("ham")
textcat.add_label("spam")We'll convert the labels in the data to the form TextCategorizer requires. For each document, we'll create a dictionary of boolean values for each class. The model is looking for these labels inside another dictionary with the key "cats".
texts = spam["text"].values
lebels = [
{"cats": {"ham": (label == "ham"), "spam": (label == "spam")}}
for label in spam["label"]
]# combine the texts and labels into a single list
data = list(zip(texts, lebels))# split train and valid data
split = int(len(data) * 0.8)
train_data, valid_data = data[:split], data[split:]# create the batch generator
spacy.util.fix_random_seed(1)
from spacy.util import minibatch
batches = minibatch(train_data, size=8)- In general it's more efficient to train models in small batches.
minibatchfunction returns a generator yielding minibatches for training.
# create an optimizer to update the model
optimizer = nlp.begin_training()
# iterate through minibatches
for batch in batches:
# split texts and labels
texts, labels = zip(*batch)
# update the model
nlp.update(texts, labels, sgd=optimizer)- Each batch is a list of (text, label) but we need to send separate lists for texts and labels to
nlp.update.
That was just one training loop (or epoch) through the data. The model will typically need multiple epochs.
optimizer = nlp.begin_training()
import random
random.seed(1)
spacy.util.fix_random_seed(1)
losses = {}
for epoch in range(10):
random.shuffle(train_data)
batches = minibatch(train_data, size=8)
for batch in batches:
texts, labels = zip(*batch)
nlp.update(texts, labels, sgd=optimizer, losses=losses)
print(losses)# split text and labels from valid_data
texts, labels = zip(*valid_data)# tokenize texts
docs = [nlp.tokenizer(text) for text in texts]We can make predictions with the predict() method.
# use TextCategorizer to predict the scores/probability for each doc
textcat = nlp.get_pipe("textcat")
scores, _ = textcat.predict(docs)# find the label with the highest score/probability
predicted_labels = scores.argmax(axis=1)# evaluate the model
correct_predictions = predicted_labels == labels
correct_predictions.mean()Note: Look at the exercise file for functional solution.
Explore an idea that ushered in a new generation of NLP techniques. #
We can usually represent the text numerically better with word embeddings. Word embeddings (also called word vectors) represent each word numerically in such a way that the vector corresponds to how that word is used or what it means. Vector encodings are learned by considering the context in which the words appear. Words that appear in similar contexts will have similar vectors. For example, vectors for "leopard", "lion", and "tiger" will be close together, while they'll be far away from "planet" and "castle".
Relations between words can be examined with mathematical operations. Subtracting the vectors for "man" and "woman" will return another vector. If you add that to the vector for "king" the result is close to the vector for "queen."
These vectors can be used as features for machine learning models. spaCy provides embeddings learned from a model called Word2Vec. We can access them by loading a large language model like en_core_web_lg. Then they will be available on tokens from the .vector attribute.
# load the large model to get the vectors
import spacy
nlp = spacy.load("en_core_web_lg")text = "These vectors can be used as features for machine learning models."
# disable other pipes, we don't need them, it'll speed up this part
import numpy as np
with nlp.disable_pipes():
vectors = np.array([token.vector for token in nlp(text)])
vectors.shape
>>> (12, 300)- These are 300-dimensional vectors, with one vector for each word.
We only have document-level labels and our models won't be able to use the word-level embeddings. So, we need a vector representation for the entire document.
There are many ways to combine all the word vectors into a single document vector we can use for model training. A simple and surprisingly effective approach is simply averaging the vectors for each word in the document. Then, we can use these document vectors for modeling. spaCy calculates the average document vector which we can get with doc.vector.
# load spam data
import pandas as pd
spam = pd.read_csv("../input/nlp-course/spam.csv")
with nlp.disable_pipes():
doc_vectors = np.array([nlp(text).vector for text in spam["text"]])
doc_vectors.shape
>>> (5572, 300)- Why 300 columns? This is the same length as word vectors. Probably something about Eigenvectors. Huh?
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
doc_vectors, spam["label"], test_size=0.1, random_state=1
)With the document vectors, we can train scikit-learn models, xgboost models, or any other standard approach to modeling. Here we'll use Support Vector Machines (SVMs). Scikit-learn provides an SVM classifier LinearSVC.
# define model
from sklearn.svm import LinearSVC
svc = LinearSVC(random_state=1, dual=False, max_iter=10000)- Setting
dual=Falsespeeds up training.
# fit and evaluate the mdoel
svc.fit(X_train, y_train)
svc.score(X_test, y_test)Documents with similar content generally have similar vectors. So we can find similar documents by measuring the similarity between the vectors. A common metric for this is the cosine similarity which measures the angle between two vectors, a and b:
This can vary between -1 and 1, corresponding complete opposite to perfect similarity, respectively. To calculate it, we can use the metric from scikit-learn or write our own function.
def cosine_similarity(a, b):
return np.dot(a, b) / np.sqrt(a.dot(a) * b.dot(b))a = nlp("REPLY NOW FOR FREE TEA").vector
b = nlp(
"According to legend, Emperor Shen Nung discovered tea when leaves from a wild tree blew into his pot of boiling water."
).vector
cosine_similarity(a, b)Sometimes people center document vectors when calculating similarities. That is, they calculate the mean vector from all documents, and they subtract this from each individual document's vector. Because sometimes our documents will already be fairly similar. For example, reviews of businesses compared to news articles, technical manuals, and recipes. We end up with all the similarities between 0.8 and 1 and no anti-similar documents (similarity < 0). When the vectors are centered, we are comparing documents within our dataset as opposed to all possible documents.
review_vec = nlp("REPLY NOW FOR FREE TEA").vector
# calculate the mean for the document vectors, should have shape (300,)
vec_mean = doc_vectors.mean(axis=0)
# center the document vectors
centered = doc_vectors - vec_mean
# calculate similarities for each document in the dataset, subtract the mean from the review vector
sims = np.array([cosine_similarity(review_vec - vec_mean, vec) for vec in centered])
# get the index for the most similar document
most_similar = sims.argmax()