Contributors:
- Ngo Nhat Khang
- Hy Truong Son (Correspondent / PI)
Papers:
- Predicting Drug-Drug Interactions using Deep Generative Models on Graphs, NeurIPS 2022 (AI for Science) https://arxiv.org/pdf/2209.09941.pdf
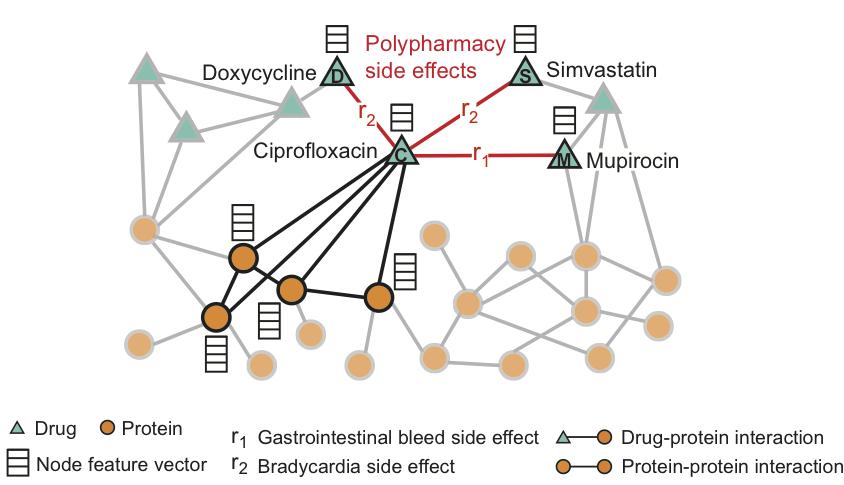
Figure taken from http://snap.stanford.edu/decagon/
- Pytorch
- Pytorch Geometric
Recommend using Conda for easy installation.
Make sure a Data folder is created in each data's subfolder. Then, you should donwload data from the links below and locate them into the Data folders as:
├── Anticancer
│ ├── Data
│ ├── ...
└── Polypharmacy
│ ├── Data
│ ├── ...
└── README.md
Download from ADRML
Download from Decagon
cd Anticancer/
bash train.sh- Train GAE
cd Polypharmacy/ python3 train_hetero_gae.py --seed 1 --num_epoch 300 --lr 1e-3 --chkpt_dir ./ --dropout 0.1 --device cuda:0 - Train VGAE
cd Polypharmacy/ python3 train_hetero_vgae.py --seed 1 --num_epoch 300 --lr 1e-3 --chkpt_dir ./ --dropout 0.1 --device cuda:0 --latent_encoder_type linear - Train VGAE + Morgan fingerprints
cd Polypharmacy/ python3 train_hetero_vgae_morgan.py --seed 1 --num_epoch 300 --lr 1e-3 --chkpt_dir ./ --dropout 0.1 --device cuda:0 --latent_encoder_type linear
@article{Zitnik2018,
title={Modeling polypharmacy side effects with graph convolutional networks},
author={Zitnik, Marinka and Agrawal, Monica and Leskovec, Jure},
journal={Bioinformatics},
volume={34},
number={13},
pages={457–466},
year={2018}
}
@article{ahmadi2020adrml,
title={ADRML: anticancer drug response prediction using manifold learning},
author={Ahmadi Moughari, Fatemeh and Eslahchi, Changiz},
journal={Scientific reports},
volume={10},
number={1},
pages={1--18},
year={2020},
publisher={Nature Publishing Group}
}