This is our implementation for the paper
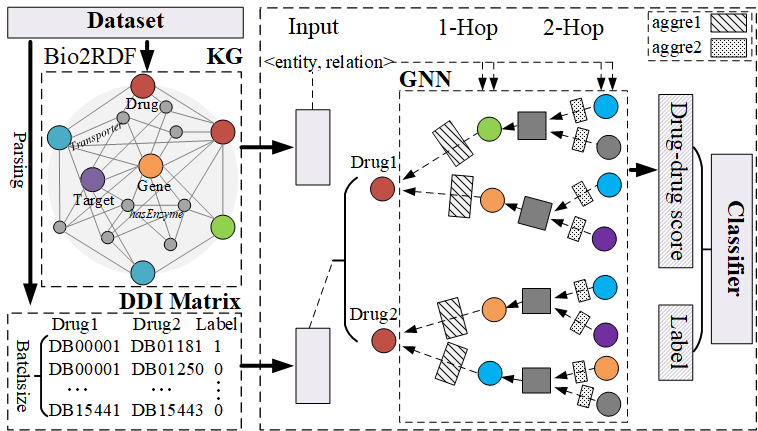
Figure 1 shows the overview of KGNN. It takes the parsed DDI matrix and knowledge graph obtained from preprocessing of dataset as the input. It outputs the interaction value for the drug-drug pair.Xuan Lin, Zhe Quan, Zhi-Jie Wang, Tengfei Ma and Xiangxiang Zeng. KGNN: Knowledge Graph Neural Network for Drug-Drug Interaction Prediction. IJCAI' 20 accepted.
To run the code, you need the following dependencies:
- Python == 3.6.6
- Keras == 2.3.0
- Tensorflow == 1.13.1
- scikit-learn == 0.22
You can create a virtual environment using conda.
conda create -n kgnn python=3.6.6
source activate kgnn
git clone https://github.com/xzenglab/KGNN.git
cd KGNN
pip install -r requirement.txt We just provide the preprocessed KG from KEGG dataset owing to the size limited. Please feel free contact us if you need the KG from DrugBank dataset (V5.1.4). The construction of KG please refer to Bio2RDF tool in detail.
python run.py(To appear)
For any clarification, comments, or suggestions please create an issue or contact Jacklin.