TODO: some documentation.
Just the code for now. A very simple implementation of a modularly trainable GP calculator and a NEB relaxation algorithm that uses it.
- Kernel construction is inspired by scikit-learn.
- GP algorithms are inspired by scikit-learn and gpmin in ASE.
- NEB relaxation is inspired by Catlearn
A very simple NEB example.
from ase.calculators.emt import EMT
import btoms
# Use ASE to make an interpolation.
images = btoms.utils.get_neb_trajectory(
'test/initial.traj',
'test/final.traj',
nimages=15)
# Kernel selection is inspired for Scikit-learn
kernel = btoms.GRBF(0.1) + btoms.GWhiteNoise(1e-3, 'fixed')
neb = btoms.NEB(
trajectory=images,
calculator=EMT(),
kernel=kernel)
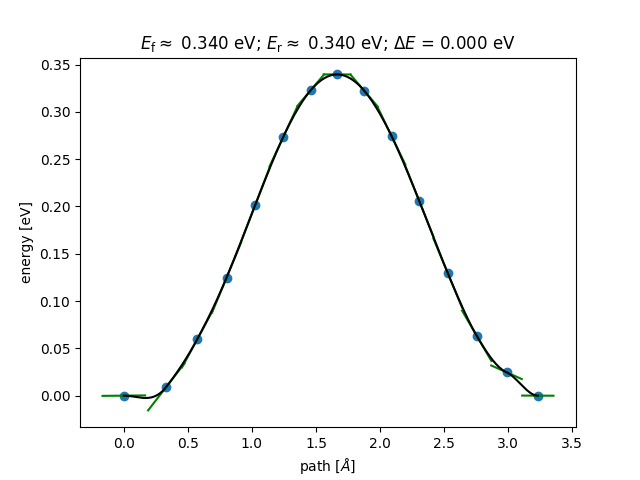
neb.run(steps=30)The example will output two files. The last predicted NEB path as neb.json and the single point calculations which were evaluated and used to train the Gaussian Process calculator in targets.json.
import matplotlib.pyplot as plt
from ase.neb import NEBTools
from ase.io import read
images = read('neb.json', ':')
neb = NEBTools(images)
neb.plot_band()
plt.savefig('test/neb.png')