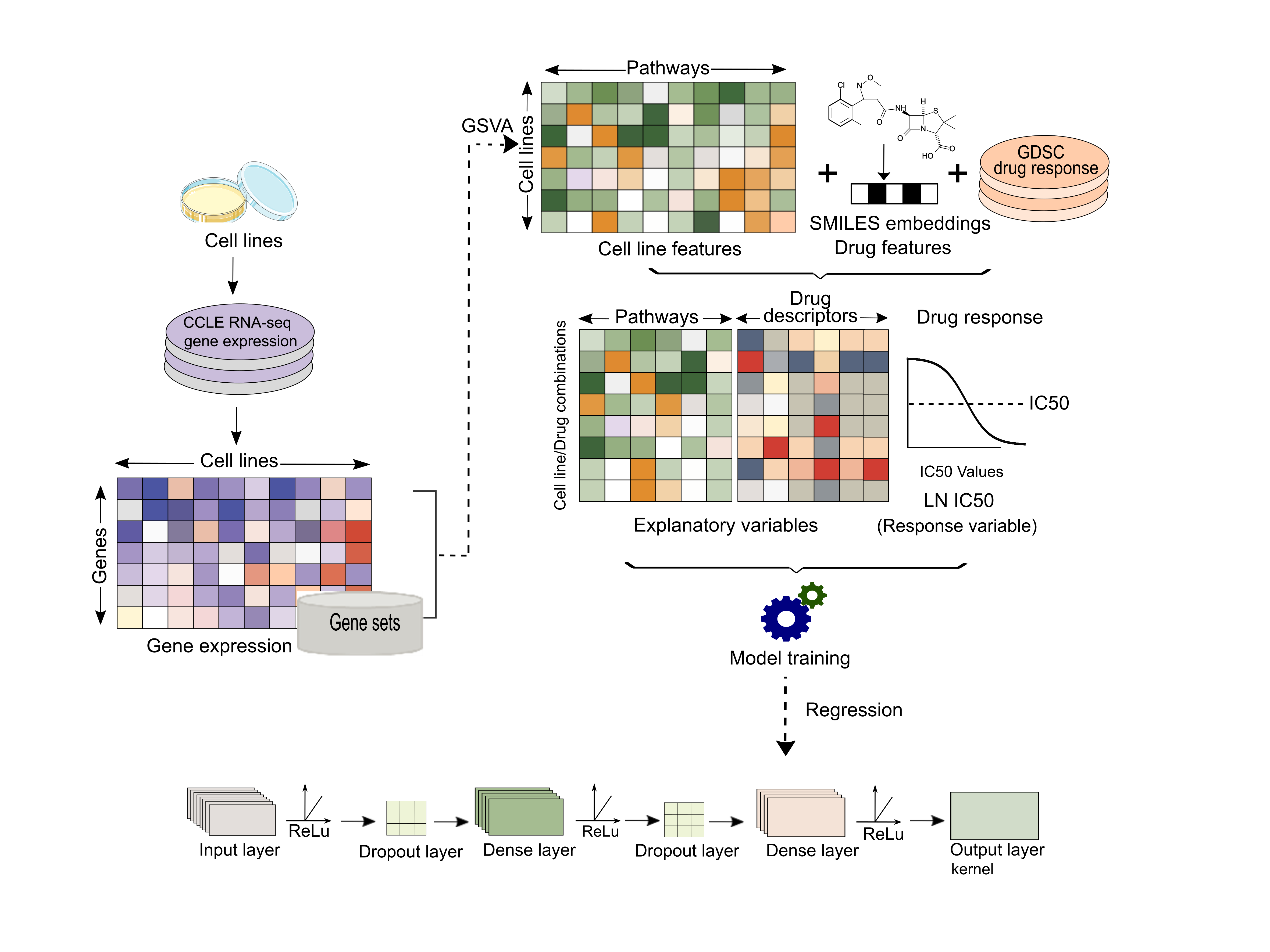
Precily, Deep neural network based framework for prediction of drug response in both in vivo and in vitro settings.
The machine learning and deep learning models were trained on a system with a following specs:
-
RAM: 16+ GB
-
CPU: 16+ cores
The expected runtime for the demo code is less than a minute. The typical installation time for setting up the environment is approximately 6 minutes. The runtimes are generated using a computer with the specs (16GB RAM, 4 cores@ 2.42 GHz)
-
Linux (Ubuntu 20.04.3)
-
Windows (version 21H2, OS Build 22000.739)
This resource provides code to reproduce key results in the manuscript.
git clone https://github.com/SmritiChawla/Precily.git
- keras 2.8.0
- keras-tuner 1.1.0
- caret(v6.0.90)
- glmnet(v4.1.3)
- ranger(v0.13.1)
- GSVA(v1.40.1)
- h2o(v3.36.0.4)
- PubChemPy(v1.0.4)
- SMILESVec
- EDASeq(v2.26.1)
- impute(v1.66.0)
- ggpubr(v0.4.0)
- ggplot2(v3.3.5)
- pheatmap(v1.0.12)
- parsnip(v0.2.1)
- ggridges(v0.5.3)
-
Data folder contains a zipped file used for training CCLE/GDSC and CCLE/CTRPv2 models. The CCLE GSVA scores are also included in this folder.
-
Data folder also contains GSVA pathway scores for Prostate cancer datasets used for evaluation of Precily. For reproducibility, respective datasets are also provided in each figure directory.
-
Use R script EnvSet.R provided in the directory EnvironmentSetup to set up an environment in R for loading python trained deep neural network models.
-
Run individual codes from the figure wise directories for reproducing manuscript results.
-
We have provided pre-trained CCLE/GDSC models for drug response prediction. DrugsPred.R is the main function used for making predictions and takes the following inputs:
1. Pathway enrichment scores/GSVA scores
2. Metadata file. This file contains drugs and cell line names, cancer types and drug descriptors
3. Cancer Type. Type of cancer to be used for the input test dataset. Cancer types are encoded in form of TCGA abbreviations.
Example code:
Predictions = drugPred(enrichment.scores,metadata,"BRCA")
The output file is a list of drug response predictions for the individual samples present in the test set.
-
Fig 1: This folder contains codes for evaluating the CCLE/GDSC data trained model and includes the following subdirectories:
Fig 1c. Pre-trained models to assess performance of Precily.
Fig 1d. CCLE/GDSC2 data trained models and independent test dataset used to evaluate Precily based deep neural network model.
Fig 1e. CCLE/CTRPv2 data trained models and independent test dataset used to evaluate Precily based deep neural network model.
-
Fig 2: This folder contains codes used for evaluating CCLE/GDSC data trained model on scRNA-seq datasets and includes the following subdirectories:
Fig 2a. This folder contains CCLE/GDSC dataset trained models, processed Kinker, G. S. et al. scRNA-seq dataset and ground truth labels for evaluation of Precily.
Fig 2b. This folder contains code for assessing the efficiency of our model on Lee et al. scRNA-seq profiles of MDA-MB-231 breast cancer cells.
-
Fig 3: This directory contains codes for evaluating Precily on the prostate cancer cell line datasets. The GSVA scores for untreated prostate cancer cell lines and treated LNCaP cell lines are provided for drug response prediction.
Fg 3a & b. Evaluation of Precily using Prostate cancer (PCa) baseline cell lines. This folder contains CCLE/GDSC2 models retrained after removing the concerned cell lines present in PCa cell line independent test set and GSVA scores.
Fig 3c. This folder contains CCLE/GDSC pre-trained models to assess the performance of Precily on LNCaP cell lines and compare it with groud truth from GDSC database.
Fig 3d. This folder contains pre-trained CCLE/GDSC models and enrichment scores of LNCaP cell lines under different treatment conditions for making predictions.
Fig 3e. This folder contains enrichment scores of LNCaP cell lines under different treatment conditions to investigate the enrichment of proliferation related pathways.
Fig 3f. This folder contains pre-trained CCLE/GDSC models and enrichment scores of LNCaP cell lines under different treatment conditions for analysis of predictions of DNA replication pathway related drugs.
Fig 3g. The folder contains CCLE/GDSC pre-trained models and file containing SMILES for Orlistat and Metformin drugs.
-
Fig 4: This directory contains codes for reproducing results for LNCaP derived xenografts datasets. We have included predictions for 155 drugs for 54 samples and GSVA scores.
Fig 4b. CCLE/GDSC pre-trained models for making predictions on PCa xenograft data. This directory also contains pre-computed predictions used for downstream analysis. Our xenograft dataset clustered into 3 groups based on the predictions.
Fig 4c. This folder contains code to visualize distribution of predictions across 3 clusters.
Fig 4d. This folder contains pathway enrichment scores of PCa xenograft dataset to visualize the distribution of proliferation related pathways across 3 clusters.
Fig 4e. This directory contains pre-computed predictions on PCa xenograft dataset for visualization of overall predictions across different tumor types.
Fig 4f. This folder contains pathway enrichment scores of PCa xenograft dataset to visualize the distribution of proliferation related pathways across tumor types.
Fig 4g. This directory contains pre-computed predictions on PCa xenograft dataset for analyzing predictions of EGFR related inhibitors across tumor types.
Fig 4h. This directory contains code and SMILES to predict drug response for three drugs (Apalutamide, Enzalutamide and Bicalutamide). These three drugs are not part of our training data.
-
Fig 5. This directory contains codes for evaluating model trained on TCGA patient RNA-seq bulk profiles.
Fig 5a. This folder contains script and data used for training AutoML models.
Fig 5b. This folder contains predictions obtained on the TCGA test dataset using the best AutoML model and code for computing survival based on these predictions.
Fig 5d-f. This folder contains codes for evaluating TCGA model using the external Wagle, Nikhil, et al. dataset.
-
Supplementary directory contains codes for reproducing supplementary figures.