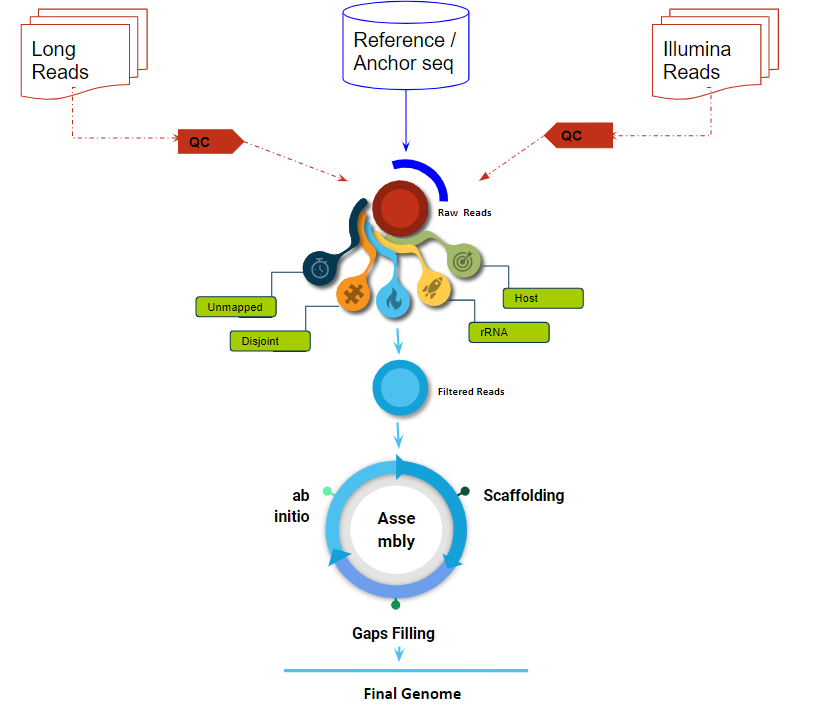
Setu: A pipeline for robust assembly of the SARS-CoV-2 genome.
+TO BE NOTED:
+We are always working to improve SETU so any bug reports, suggestions and general feedback would be highly welcome. Setu (sanskrit सेतु) means bridge. It bridges all the reads to genome.
______ _____/ |_ __ __
/ ___// __ \ __\ | \
\___ \\ ___/| | | | /
/____ >\___ >__| |____/ v0.2
\/ \/ bridging the SARS-CoV-2 genome
Clone the repository using git:
git clone https://github.com/jnarayan81/setu.git
Required dependencies can be installed in a separate conda environment named setu through:
cd setu
conda create -f env_setu.yml
Setu requires the following dependencies to be installed:
- Python 3.7
- Trimmomatic
- BWA-MEM
- Samtools
- Bedtools
- Spades
- Ragout
- QUAST
- R >=3.6.0
- Reshape package
Alternatively, all dependencies can be installed through Conda.
Setu supports only paired-end Illumina reads at the moment, work on long-reads, command is as follows:
Paired-end reads:
./setu.sh -k yes -m pe -t 1 -r paired_1.fastq,paired_2.fastq -f on -o OutputDirectory
Please note that there's no space after the comma when specifying reads using the -r flag.
Assembly of long-reads and hybrid-reads is currently ongoing and will be updated.
You can test your installation by running:
./test_run.sh
Setu: A pipeline for robust assembly of the SARS-CoV-2 genome. It has three mode of genome assembly: 1. Paired-End 2. Hybrid 3. Long reads.
The promise of setu:
- Implement recent NGS techniques to achieve reliable genome assembly.
- Maintain flexibility in reads type selection.
- Build on standard Conda and Python packages.
In a nutshell, this pipeline is intended to use all types of NGS reads to generate a genome of high quality.
Consult Jitendra Narayan at jnarayan81@gmail.com or info@bioinformaticsonline.com for any support.
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a new Betacoronavirus strain that infects humans. This disease is the cause of the ongoing Coronavirus Disease (CoViD2019) pandemic. Because of the rapid innovation and decreasing prices of high throughput sequencing technologies, the virus has been sequenced internationally in a large number of people who have been infected. While next-generation sequencing (NGS) technology provides a reliable method of identifying potential infections in clinical specimens, simple and user-friendly bioinformatics workflows are necessary to acquire a complete viral genome sequence with the greatest accuracy. We have developed a thorough workflow for evaluating and decoding SARS-CoV 2 sequencing data using open source technologies. It entails complete sequence elimination of host- or bacteria-related NGS reads prior to de novo assembly, resulting in the quick and accurate assembly of viral genome metagenomic sequences.
Illustrating procedures for assembly of the SARS-CoV-2 genome.
June 1, 2022: Release v0.2, see release notes here
- June 2020: CoViD Assembler
If you use setu in your research, please cite us as follows:
Nityendra Shukla¹, Neha Srivastava³, Prachi Srivastava³*, Jitendra Narayan¹* Setu: A Pipeline for the robust Assembling of the SARS-CoV-2 Genome https://github.com/jnarayan81/setu, 2023. Version 0.2
BibTex:
@misc{setu,
author={Nityendra Shukla¹, Neha Srivastava³, Prachi Srivastava³*, Jitendra Narayan¹*}
title={{Setu}: {A Pipeline for the robust Assembling of the SARS-CoV-2 Genome}},
howpublished={https://github.com/jnarayan81/setu},
note={Version 0.2},
year={2023}
}
This project welcomes contributions and suggestions.
For more information contact jnarayan81@gmail.com or (mailto:info@bioinformaticsonline.com) with any additional questions or comments.