Software for high density electrophysiology.
- 25/11/17: We are currently developing a new implementation of these methods. Speed is much improved (7kHz from 4,096 channels can be analysed in real-time), and other dense probes such as Neuropixel are supported (with better than real-time performance). If you would like to try it, get in touch with us.
-
The spike sorting method is now published in a paper in Cell Reports.
-
There is now an implementations of the detection methods in python/C++, which can easily be adjusted to read any raw data file format. Code is currently provided to read the hdf5-based format used by BrainwaveX. Performance is substantially improved compare to the original C# implementations. This improvement was done by Albert Puente Encinas.
- onlineDetection: Very fast spike detection, done independently on each recording channel.
- interpolatingDetection: Spike detection with spatial interpolation. Returns cut-outs for detected events from multiple channels, which allows performing spike localisation.
- postProcessing: Programs for removing duplicate events in spikes detected with the online method, and to spatially localise spikes detected with the interpolation method.
- clustering: Perform spike sorting by location and PCA on interpolated data.
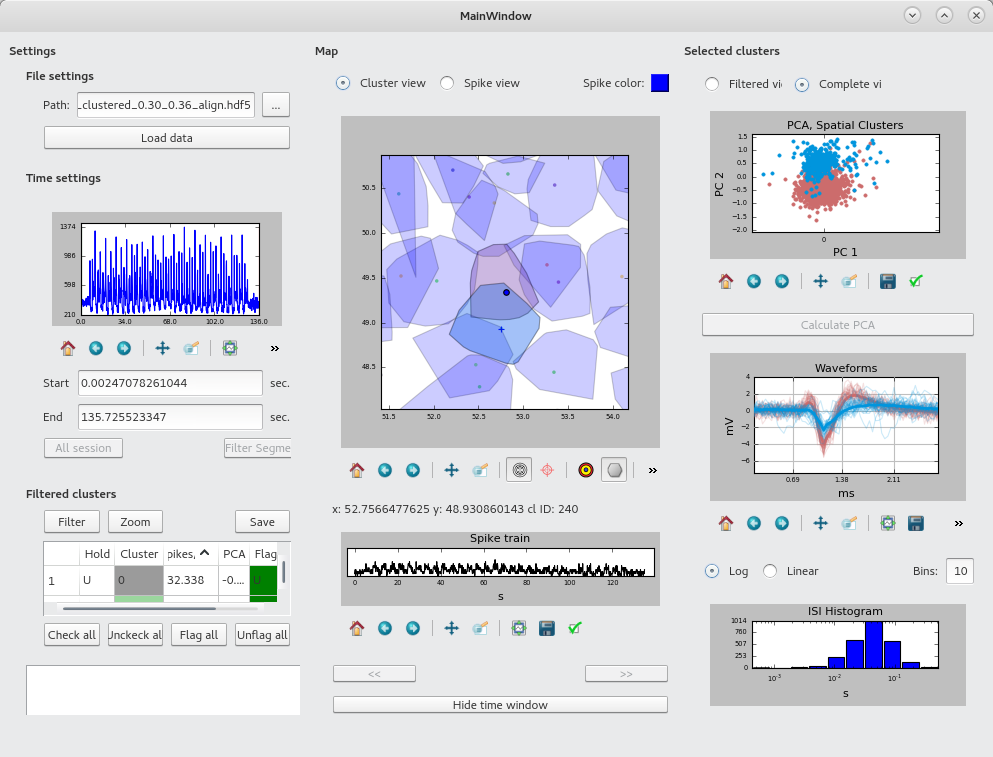
- visualisationtool: A basic GUI tool for visualising and annotating sorted spikes.
If you are interested in using this software and have questions or problems, get in touch with us. All code is released under GPL-2.0.
The project onlineDetection provides an efficient and reliable algorithm for detecting spikes in single channels. Main features are a robust noise estimate based on signal percentiles rather than moments, and fast integer based computations allowing real-time performance even when recording 1000s of channels simultaneously. While originally developed for high density multielectrode arrays, we expect this to perform well on most extracellular recordings. Peak performance is 6.5 x real time for 4,096 channels (7kHz).
This process is fully automated, as manual inspection gets very time consuming for large-scale recordings. The following steps lead all the way from raw data to sorted units:
-
Spike detection and spatial signal interpolation - currently runs in 1/4 x real time for 4096 channels, scales linearly with recording duration. Code is in the sub-project interpolatingDetection.
-
Spatial event localisation, now has about real-time performance (scales linearly with event number). The relevant code is in the sub-project postProcessing.
-
Clustering, code in clustering. This step is parallelised and extremely efficient, typical experiments with millions of spikes are clustered in minutes. The code also includes functions for noise removal and quality control.
-
Quality control, using functionality provided by the code in clustering. A full mixture model is fit to all identified units and those in their neighbourhood, to obtain an estimate of false positives and negatives. Based on these quantities, units can be rejected as poorly sorted.
These methods have for far been tested with data recorded with the 3Brain Biocam, and we are working on different data sets, including in vivo probes. Steps 1 and 2 have to be adjusted for other systems, steps 3 and 4 are already rather generic.
To run the graphical interface to inspect the results of the clustering, run
python Launcher.py
in the subfolder visualisationtool. The program reads files generated by the clustering method.
- Matthias Hennig: Spike sorting
- Oliver Muthmann: Spike detection and localisation
- Albert Puente Encinas: C++ implementation, optimisation and parallelisation
- Martino Sorbaro: Spike sorting
- Cesar Juarez Ramirez: Visualisation toolkit
J.-O. Muthmann, H. Amin, E. Sernagor, A. Maccione, D. Panas, L. Berdondini, U.S. Bhalla, M.H. Hennig MH (2015). Spike detection for large neural populations using high density multielectrode arrays. Front. Neuroinform. 9:28. doi: 10.3389/fninf.2015.00028.
G. Hilgen, M. Sorbaro, S. Pirmoradian, J.-O. Muthmann, I. Kepiro, S. Ullo, C. Juarez Ramirez, A. Puente Encinas, A. Maccione, L. Berdondini, V. Murino, D. Sona, F. Cella Zanacchi, E. Sernagor, M.H. Hennig (2016). Unsupervised spike sorting for large scale, high density multielectrode arrays. Cell Reports 18, 2521–2532. bioRxiv doi: http://dx.doi.org/10.1101/048645.