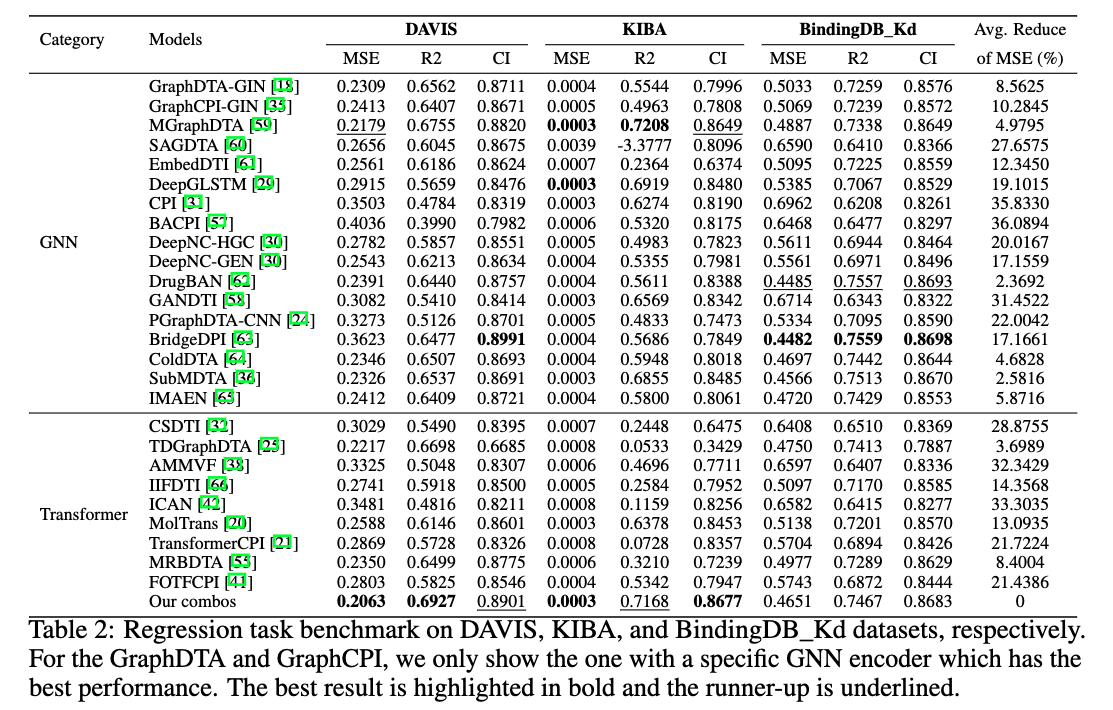
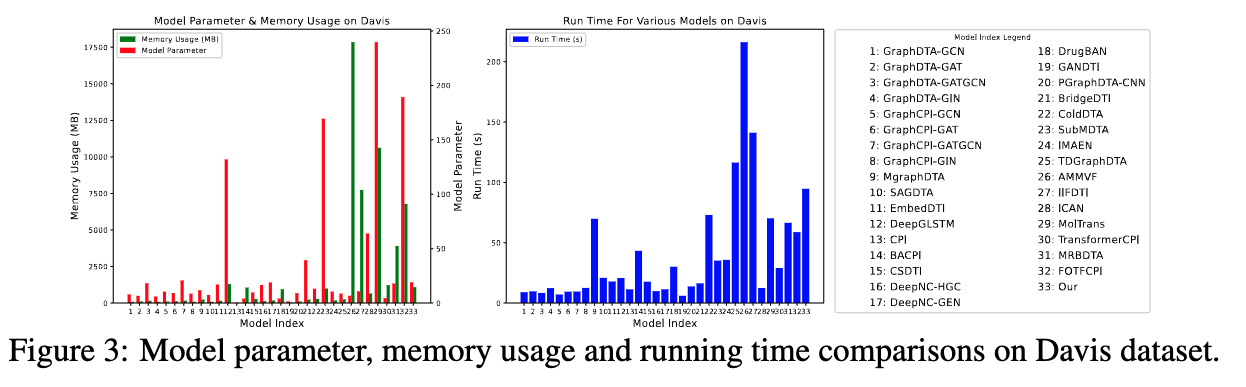
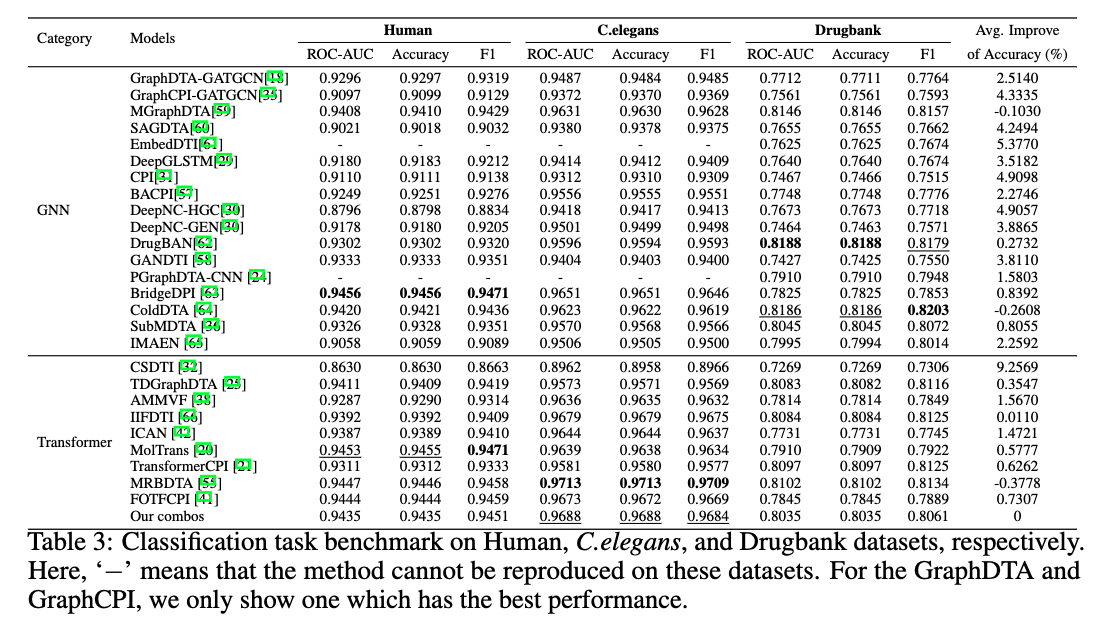
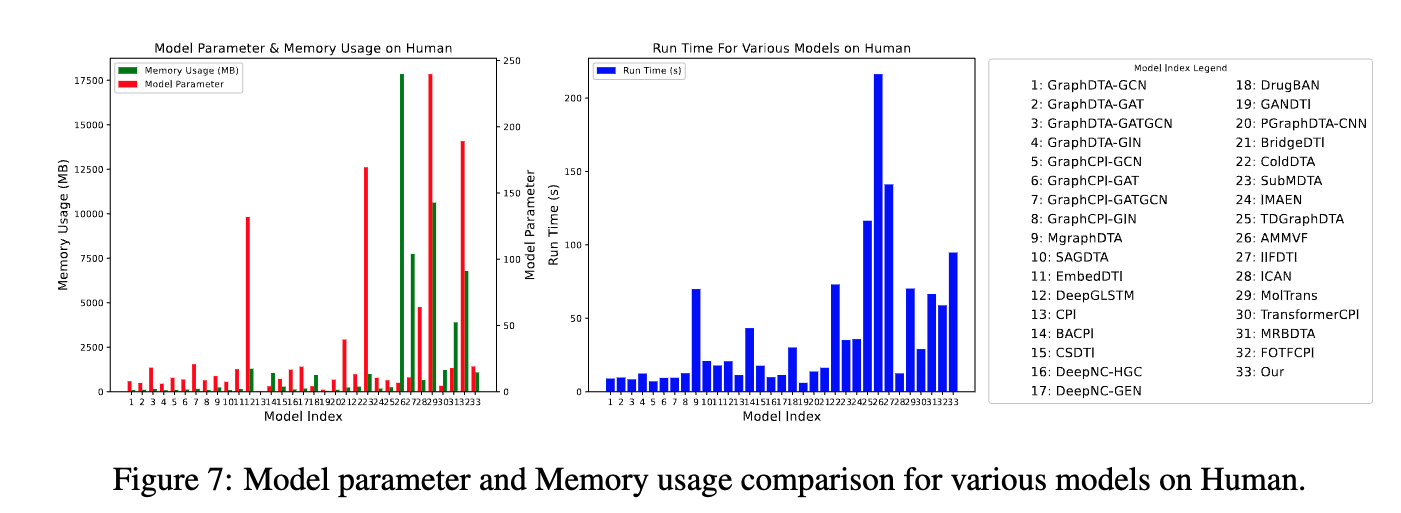
This is the official codebase of the paper Benchmark on Drug Target Interaction Modeling from a Structure Perspective.
GTB-DTI is a comprehensive benchmark customized for GNN and Transformer-based methodologies for DTI prediction.
Before you begin, you can install the required libraries using:
First, clone the repository to your local machine:
git clone https://github.com/justinwjl/GTB-DTI.git
cd your-repopython main.py -c config/model.yamlThe config.yaml file contains all the configurable parameters for training the model. You can edit this file to adjust parameters such as learning rate, batch size, and number of epochs.
Set the 'train' to 'memory_test' in the config.yaml file
task:
class: regression
model:
class: model
param:
train: memory_testThis codebase is released under the MIT License as in the LICENSE file.
If you find this paper or code helpful in your research, please cite the following paper.
@misc{zhang2024benchmarkdrugtargetinteraction,
title={Benchmark on Drug Target Interaction Modeling from a Structure Perspective},
author={Xinnan Zhang and Jialin Wu and Junyi Xie and Tianlong Chen and Kaixiong Zhou},
year={2024},
eprint={2407.04055},
archivePrefix={arXiv},
primaryClass={q-bio.QM},
url={https://arxiv.org/abs/2407.04055},
}