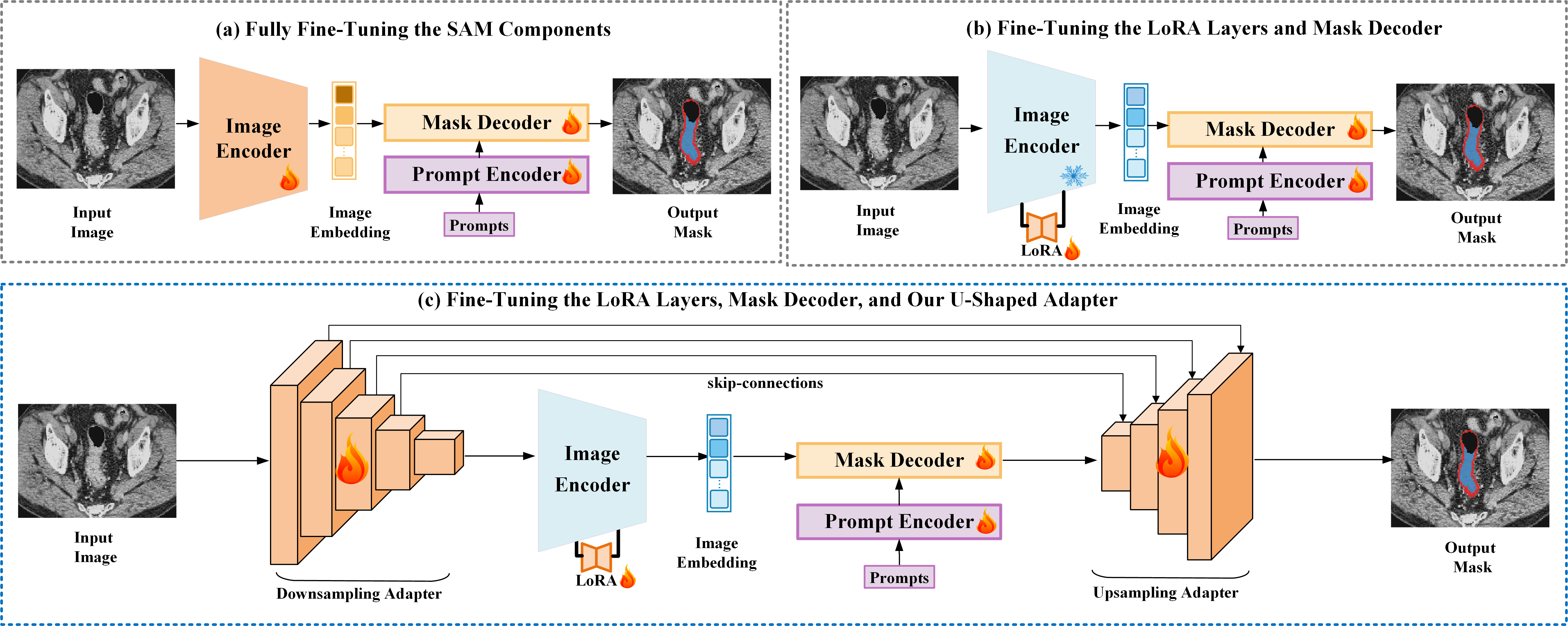
Tuning Vision Foundation Models for Rectal Cancer Segmentation from CT Scans: Development and Validation of U-SAM
- We provide our implementation of U-SAM.
- The dataloaders of CARE and WORD are also available in dataset.
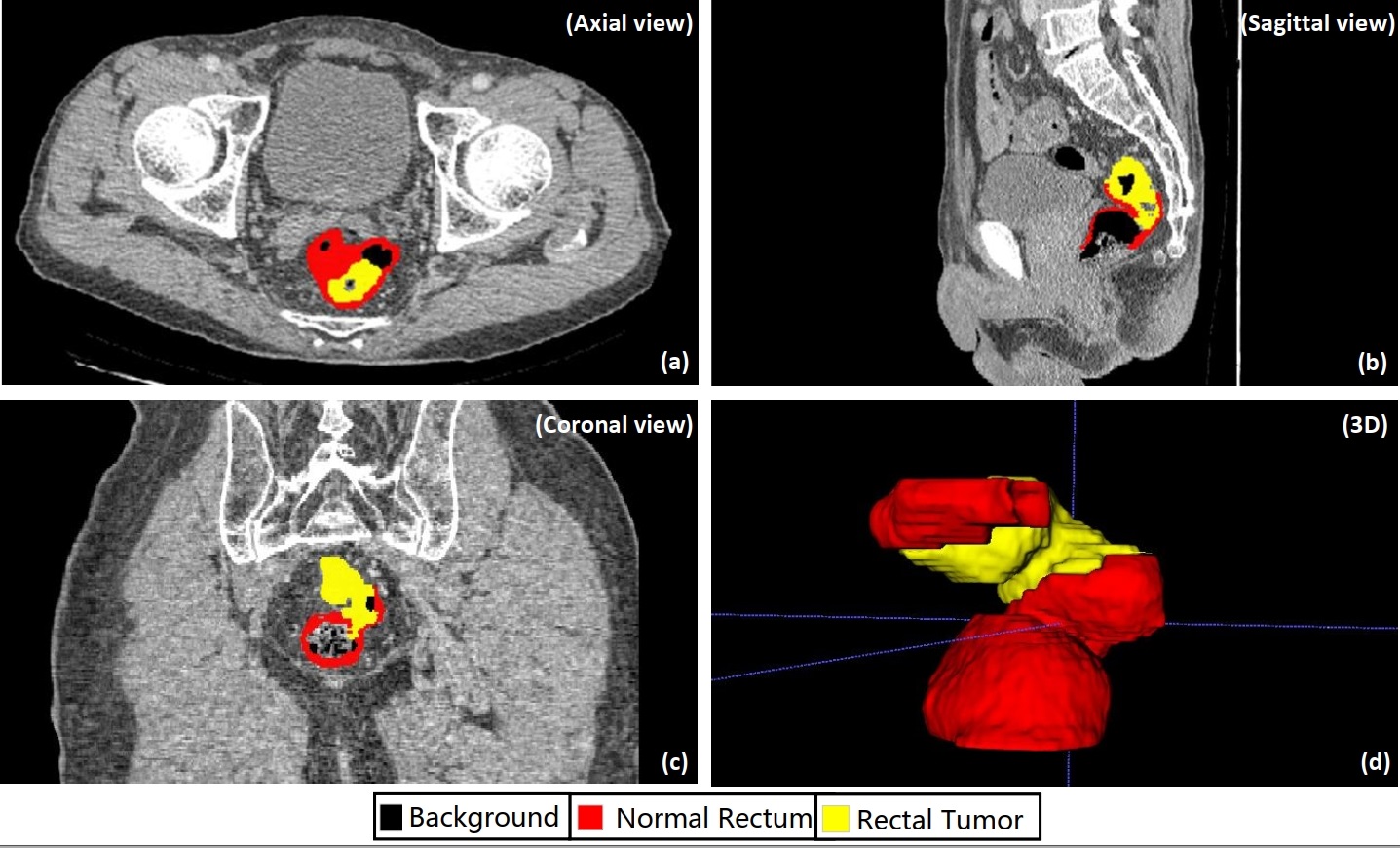
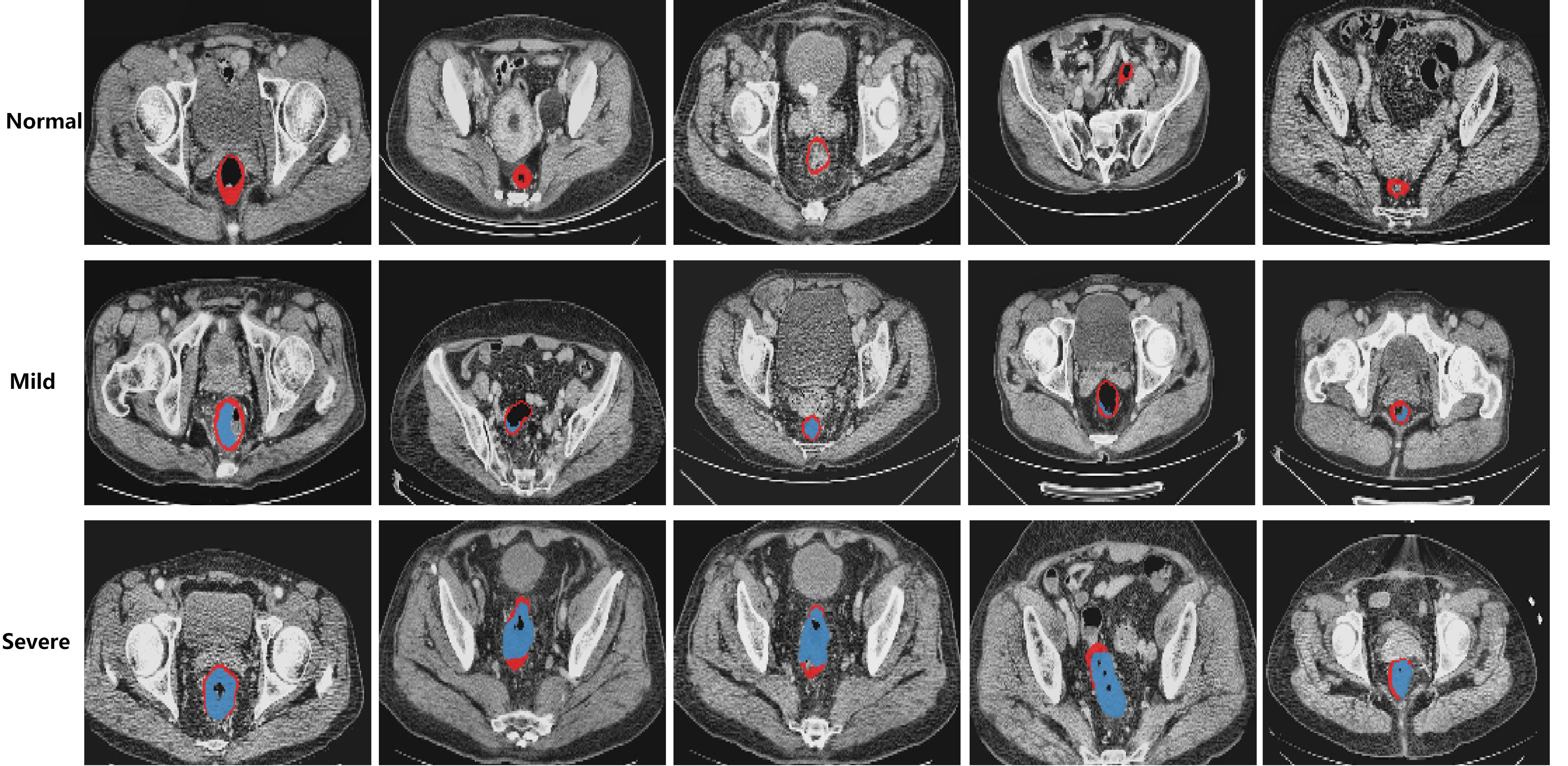
The following pictures are demonstrations of CARE.
We conducted our experiments on CARE and WORD. Here we provide public access to these datasets.
-
python==3.9.12
-
torch==1.11.0
-
torchvision==0.12.0
-
numpy==1.21.5
-
matplotlib==3.5.2
We utilized the SAM-ViT-B in our model, the pre-trained weights are supposed to be placed in the folder weight.
Pre-trained weights are available here, or you can directly download them via the following link.
vit_h: ViT-H SAM model.vit_l: ViT-L SAM model.vit_b: ViT-B SAM model.
Train 100 epochs on CARE with one single GPU:
python u-sam.py --epochs 100 --batch_size 24 --dataset rectumTrain 100 epochs on CARE with multiple GPUs (via DDP, on 8 GPUs for example):
CUDA_LAUNCH_BLOCKING=1;PYTHONUNBUFFERED=1;CUDA_VISIBLE_DEVICES=0,1,2,3,4,5,6,7 \
python -m torch.distributed.launch \
--master_port 29666 \
--nproc_per_node=8 \
--use_env u-sam.py \
--num_workers 4 \
--epochs 100 \
--batch_size 24 \
--dataset rectumFor convenience, you can use our default bash file:
bash train_sam.shEvaluate on CARE with one single GPU:
python u-sam.py --dataset rectum --eval --resume chkpt/best.pthThe model checkpoint for evaluation should be specified via --resume.
For further questions, pls feel free to contact Hantao Zhang.
Our code is based on Segment Anything and SAMed. Thanks them for releasing their codes.
If this code is helpful for your study, please cite:
@article{zhang2023care,
title={CARE: A Large Scale CT Image Dataset and Clinical Applicable Benchmark Model for Rectal Cancer Segmentation},
author={Zhang, Hantao and Guo, Weidong and Qiu, Chenyang and Wan, Shouhong and Zou, Bingbing and Wang, Wanqin and Jin, Peiquan},
journal={arXiv preprint arXiv:2308.08283},
year={2023}
}