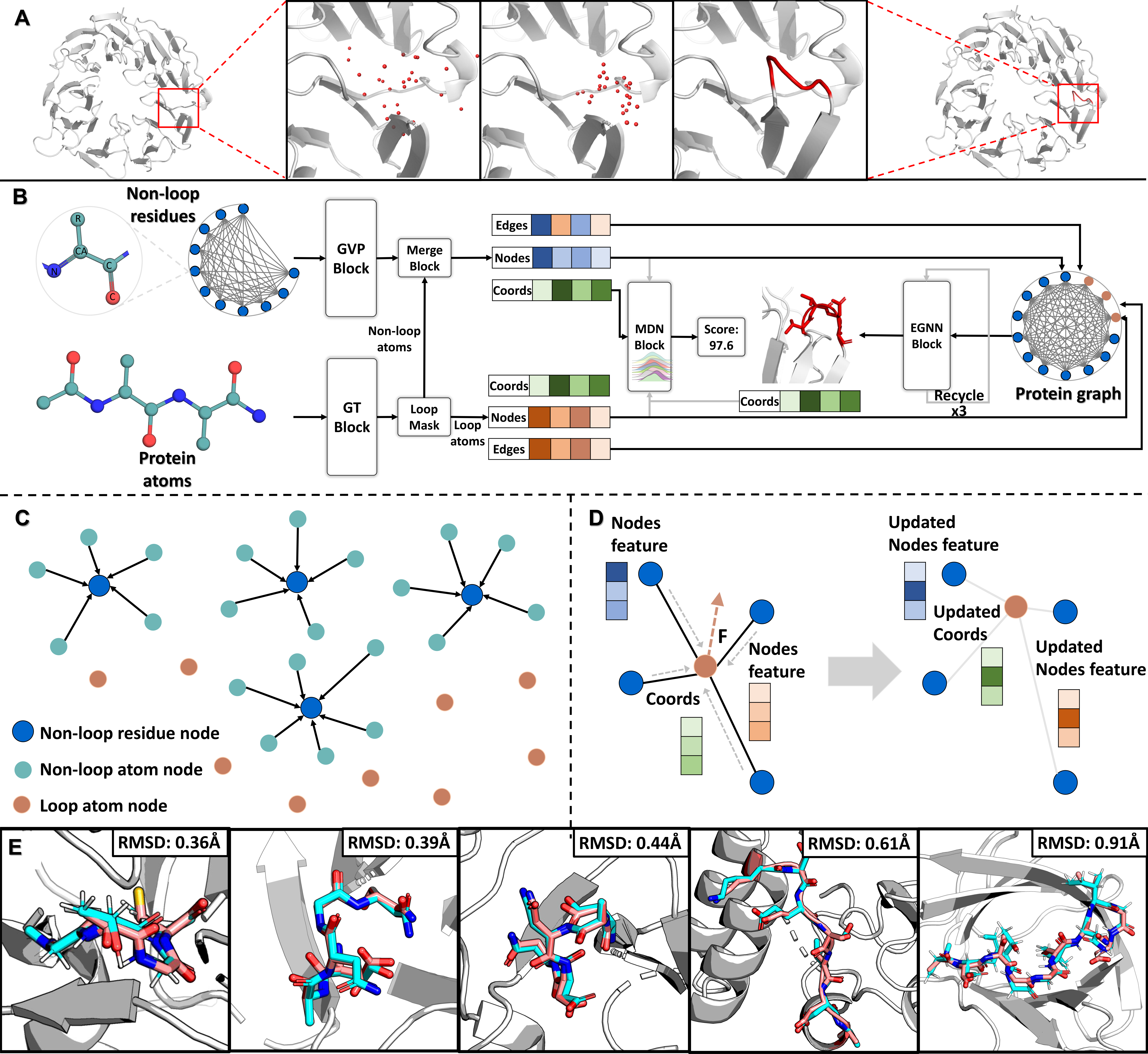
Highly accurate and efficient deep learning paradigm for full-atom protein loop modeling with KarmaLoop
KarmaLoop is novel deep learning paradigm for fast and accurate full-atom protein loop modeling.
The framework consists of four main steps: creating Python environments, selecting pocket, generating graphs and loop modeling.
You can create a new environment by the following command
conda env create -f karmaloop_env.yml -n karmaloop
conda activate karmaloop
or you can download the conda-packed file, and then unzip it in ${anaconda install dir}/envs
${anaconda install dir} represents the dir where the anaconda is installed. For me, ${anaconda install dir}=/root/anaconda3 .
mkdir /root/anaconda3/envs/karmaloop
tar -xzvf karmaloop.tar.gz -C /root/anaconda3/envs/karmaloop
conda activate karmaloop
Assume that the project is at /root and therefore the project path is /root/KarmaLoop.
We provide a shell script to reproduce the loop modeling result reported in the manuscript. If you only want to reproduce the result, simply run the following command:
cd /root/KarmaLoop
# modify the reproduciton.sh file to ensure the ${project_dir} is your project path, default is /root/KarmaLoop
bash reproduction.sh
If you wonder how to use this tool, follow the steps below.
The purpose of this step is to identify residues that are within a 12Å radius of any loop atom and use them as the pocket of the protein. The pocket file ({PDBID}_{ChainID}_{LoopStartResIdx}_{LoopEndResIdx}_12A.pdb) will also be saved on the pockets_dir.
cd /root/KarmaLoop/utils
python -u pre_processing.py
--protein_file_dir ~/your/raw/pdb/path
--pocket_file_dir ~/dst/pocket/path
--csv_file ~/your/raw/csv/path/
Example for the single demo:
cd /root/KarmaLoop/utils
python -u pre_processing.py --protein_file_dir /root/KarmaLoop/example/single_example/raw --pocket_file_dir /root/KarmaLoop/example/single_example/pocket --csv_file /root/KarmaLoop/example/single_example/single_example.csv
Example for CASP15:
cd /root/KarmaLoop/utils
python -u pre_processing.py --protein_file_dir /root/KarmaLoop/example/CASP15/raw --pocket_file_dir /root/KarmaLoop/example/CASP15/pocket --csv_file /root/KarmaLoop/example/CASP15/CASP15.csv
This step will generate graphs for protein-loop complexes and save them (*.dgl) to graph_file_dir.
cd /root/KarmaLoop/utils
python -u generate_graph.py
--protein_file_dir ~/your/raw/pdb/path
--pocket_file_dir ~/generated/pocket/path
--graph_file_dir ~/the/directory/for/saving/graph
Example for the single demo:
cd /root/KarmaLoop/utils
python -u generate_graph.py --protein_file_dir /root/KarmaLoop/example/single_example/raw --pocket_file_dir /root/KarmaLoop/example/single_example/pocket --graph_file_dir /root/KarmaLoop/example/single_example/graph
Example for CASP15:
cd /root/KarmaLoop/utils
python -u generate_graph.py --protein_file_dir /root/KarmaLoop/example/CASP15/raw --pocket_file_dir /root/KarmaLoop/example/CASP15/pocket --graph_file_dir /root/KarmaLoop/example/CASP15/graph
This step will perform loop modelling based on the graphs. (finished in about several minutes)
cd /root/KarmaLoop/utils
python -u loop_generating.py
--graph_file_dir ~/the/directory/for/saving/graph
--out_dir ~/path/for/recording/loop_conformation & confidence score
--multi_conformation whether predict miltiple loop conformations
--save_file whether save predicted loop conformations
--scoring whether calculate confidence score
--batch_size 64
--random_seed 2023
Example for the single demo:
cd /root/KarmaLoop/utils
python -u loop_generating.py --graph_file_dir /root/KarmaLoop/example/single_example/graph --out_dir /root/KarmaLoop/example/single_example/test_result --batch_size 64 --random_seed 2023 --multi_conformation --scoring --save_file
Example for CASP15:
cd /root/KarmaLoop/utils
python -u loop_generating.py --graph_file_dir /root/KarmaLoop/example/CASP15/graph --model_file /root/KarmaLoop/model_pkls/karmaloop.pkl --out_dir /root/KarmaLoop/example/CASP15/test_result --batch_size 64 --random_seed 2023 --scoring --save_file
Using OpenMM to optimize the predicted conformations of protein loop, KarmaLoop modeled loop file should be taken as input.
It may occurs 'There is no registered Platform called "CUDA"' error you can try conda install -c omnia openmm cudatoolkit=YOUR_CUDA_VERSION
to fix it. For me, the CUDA_VERSION is 11.3. It works for me.
cd /root/KarmaLoop/utils
python -u post_processing.py
--modeled_loop_dir ~/path/for/recording/loop_conformation
--output_dir ~/path/for/recording/post-processing/loop_conformation
Example for the single demo:
cd /root/KarmaLoop/utils
python -u post_processing.py --modeled_loop_dir /root/KarmaLoop/example/single_example/test_result/0 --output_dir /root/KarmaLoop/example/single_example/test_result/post
Example for CASP15:
cd /root/KarmaLoop/utils
python -u post_processing.py --modeled_loop_dir /root/KarmaLoop/example/CASP15/test_result/0 --output_dir /root/KarmaLoop/example/CASP15/test_result/post
\