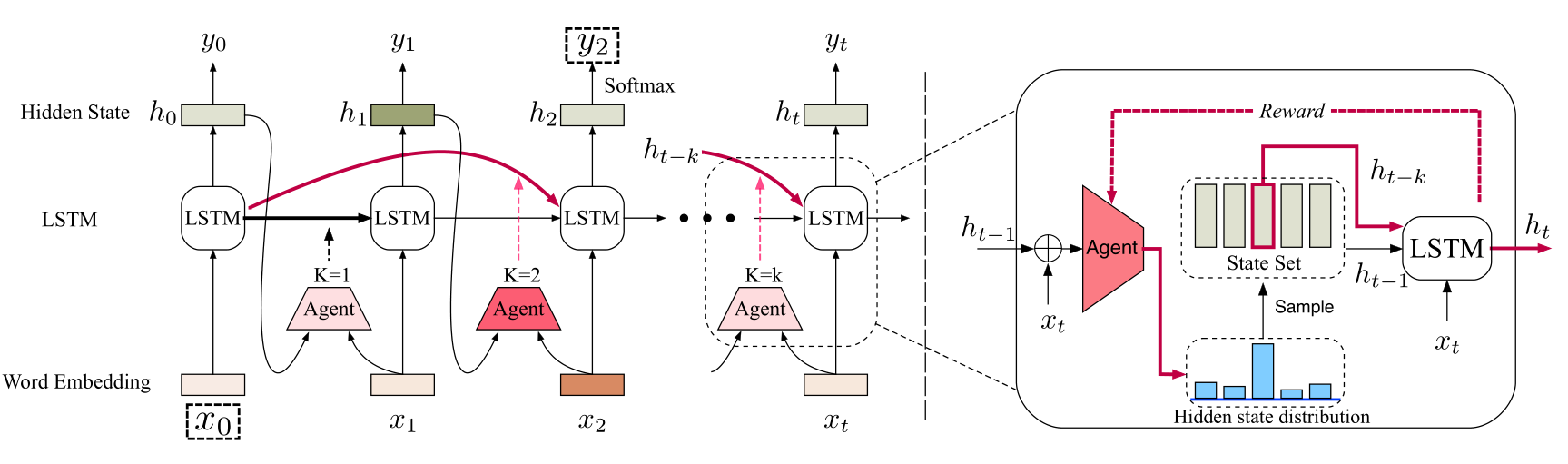
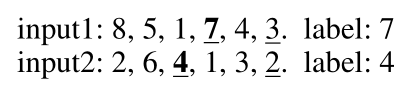TensorFlow implementation of Long Short-Term Memory with Dynamic Skip Connections (AAAI 2019).
The code is partially referred to https://github.com/tensorflow/models.
- Python 2.7 or higher
- Numpy
- TensorFlow 1.0
To evaluate the model on the Penn Treebank language model corpus, you can run the following command under the directory of ./LanguageModeling.
$ python RL_train.py --rnn_size 650 --rnn_layers 2 --lamda 1 --n_actions 5
The hyper-parameters in this model are:
| Parameter | Meaning | Default |
|---|---|---|
| rnn_size | size of LSTM internal state | 650 |
| rnn_layers | number of layers in the LSTM | 2 |
| dropout | dropout rate | 0.5 |
| lamda | lambda value of RL | 1 |
| learning_rate | initial learning rate | 1.0 |
| lr_threshold | learning rate stops decreasing after threshold | 0.005 |
| param_init | initialize parameter range of weights | 0.05 |
| n_actions | number of actions to take | 5 |
You can change these values by passing the arguments explicitly.
Given a sequence of L positive integers
To investigate this task of number prediction, you should produce datasets first under the directory of ./NumberPrediction by running:
$ python produce_data.py
Then, you can reproduce the results on number prediction dataset (with sequence length = 10+1) by executing:
$ python train_RL10_1.py
If you find this code useful, please cite us in your work:
@article{gui2018long,
title={Long Short-Term Memory with Dynamic Skip Connections},
author={Gui, Tao and Zhang, Qi and Zhao, Lujun and Lin, Yaosong and Peng, Minlong and Gong, Jingjing and Huang, Xuanjing},
journal={arXiv preprint arXiv:1811.03873},
year={2018}
}