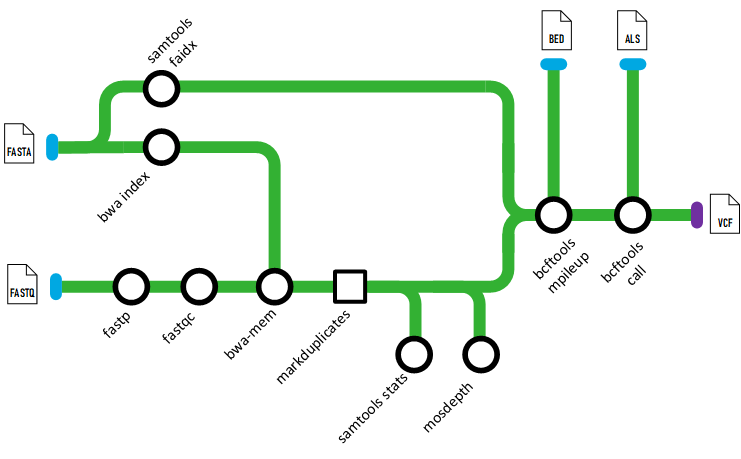
tetris is a nextflow pipeline for processing short read DNA sequencing data and calling variants.
It trims reads with (fastp), aligns with (BWA-MEM), marks duplicates (optional) with (GATK MarkDuplicates), and calls variants with (BCFTOOLS). Additionally QC stats are computed with (FastQC), (Samtools) and (mosdepth) which is aggregated into a report by (MultiQC)
The pipeline expects a CSV samplesheet as input, which should contain the sample name, sequence id, read type (single/paired) and the path to the read1 fastq file and path to the read2 fastq file (optional). It should look something similar to:
name,seqid,seq_type,fastq_1,fastq_2
PI604780,SRR17781753,paired,./docs/example_data/SRR17781753_chr1-2_R1.fastq.gz,./docs/example_data/SRR17781753_chr1-2_R2.fastq.gz
PI604779,SRR17781754,paired,./docs/example_data/SRR17781754_chr1-2_R1.fastq.gz,./docs/example_data/SRR17781754_chr1-2_R2.fastq.gz
Note the column names are important
Multiple entries can have the same sample name, however seqid must be unique
example usage and flags to be added.
- add in example data (current example data is too large for GitHub)
- ensure compatibility with single end read data
- add more optional samplesheet info to include in read group header
tetris (the nf pipeline, not the game) was originally written by LWPembleton.
A lot of inspiration and structure was taken from the Nextflow documentation, the fantastic nf-core community and modules.
Nextflow enables reproducible computational workflows.
Paolo Di Tommaso, Maria Chatzou, Evan Floden, Pablo Prieto Barja, Emilio Palumbo & Cedric Notredame.
P. Di Tommaso, et al. Nextflow enables reproducible computational workflows. Nature Biotechnology 35, 316–319 (2017) doi:10.1038/nbt.3820
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.