flowerMD is a modular “wrapper” package for molecular dynamics (MD) simulation pipeline development, designed to enable fast, reproducible, end-to- end simulation workflows with minimal user effort. This package is a wrapper for MoSDeF packages and Hoomd-Blue with a focus on simulating soft matter systems.
An object-oriented design makes flowerMD extensible and highly flexible. This is bolstered by a library-based approach to system initialization, making flowerMD agnostic to system identity, forcefield, and thermodynamic ensemble, and allowing for growth on an as-needed basis.
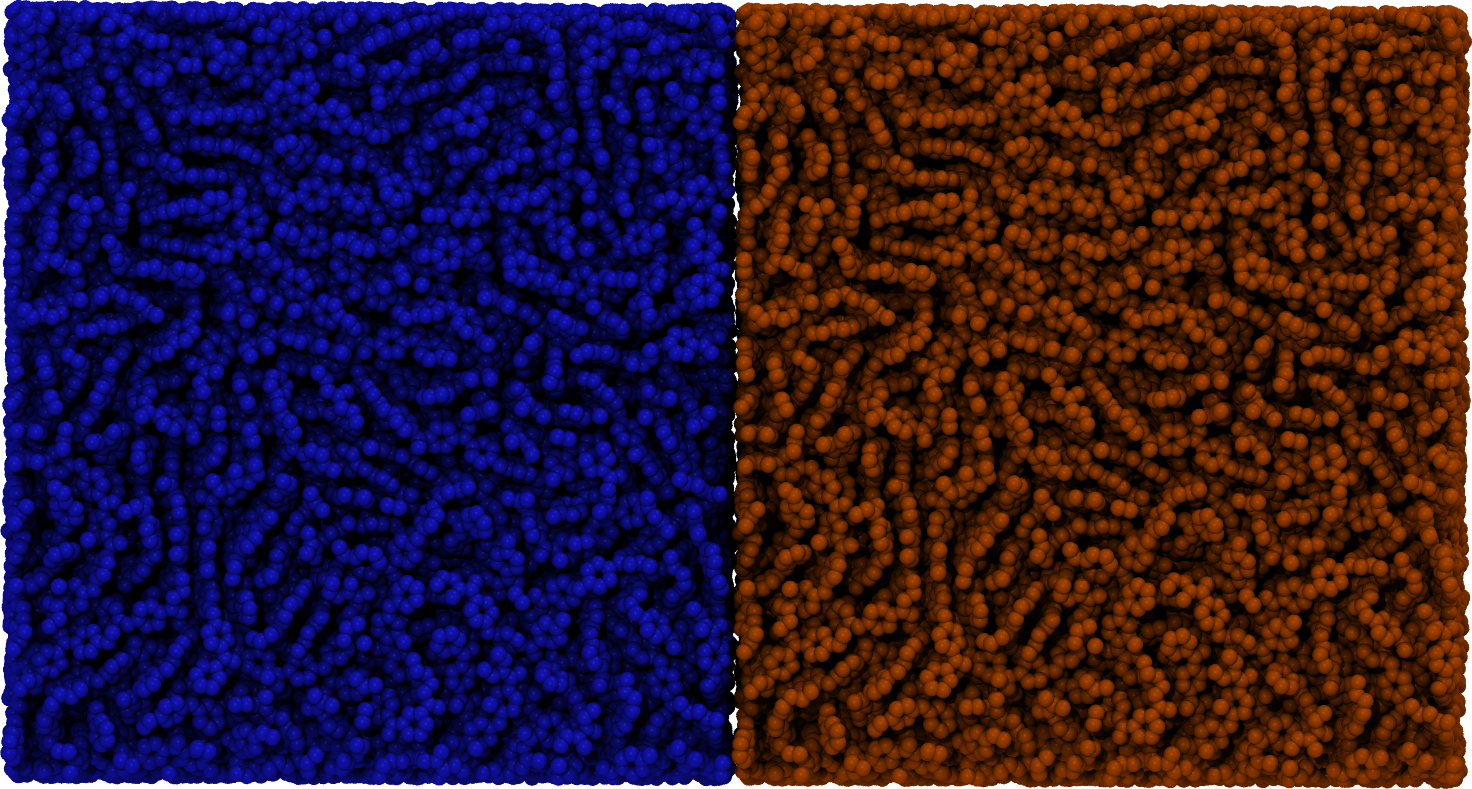
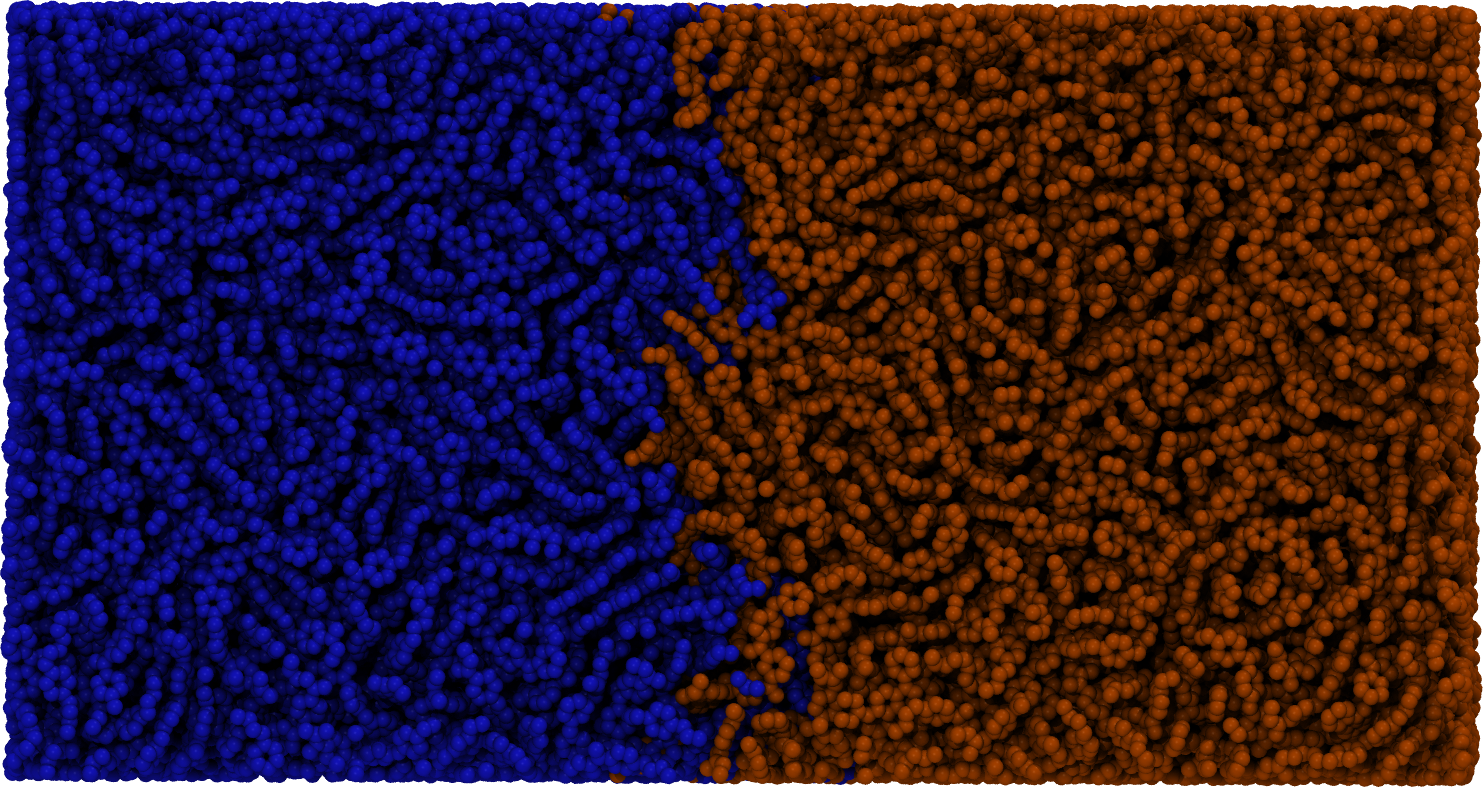
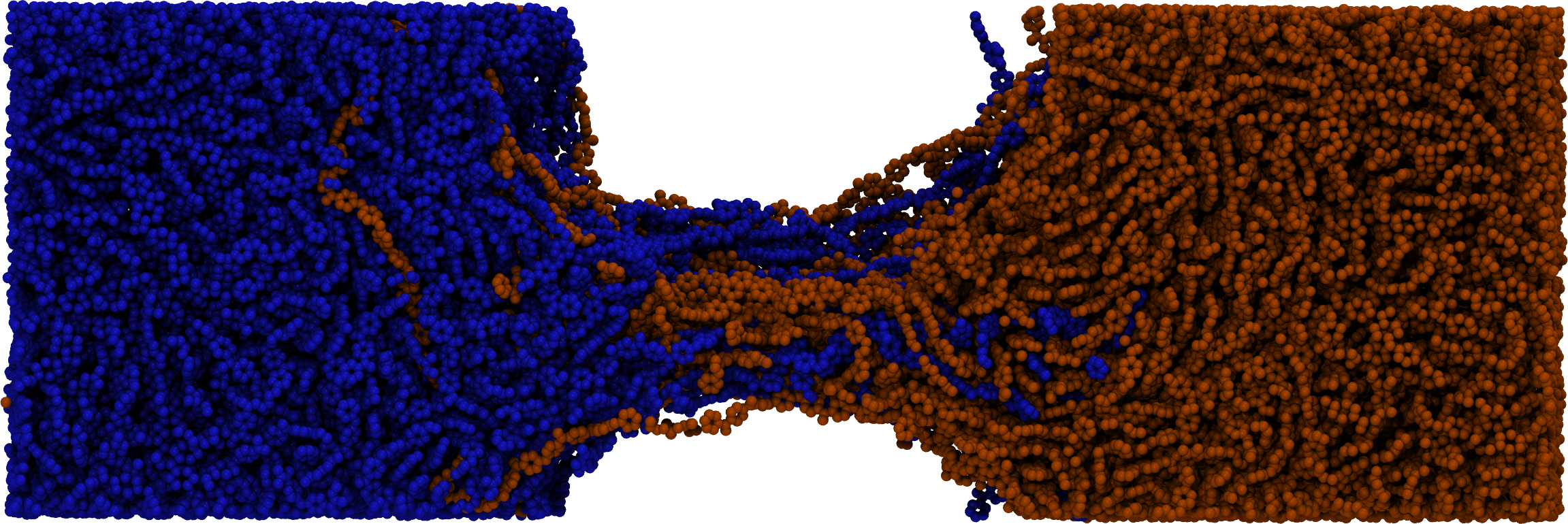
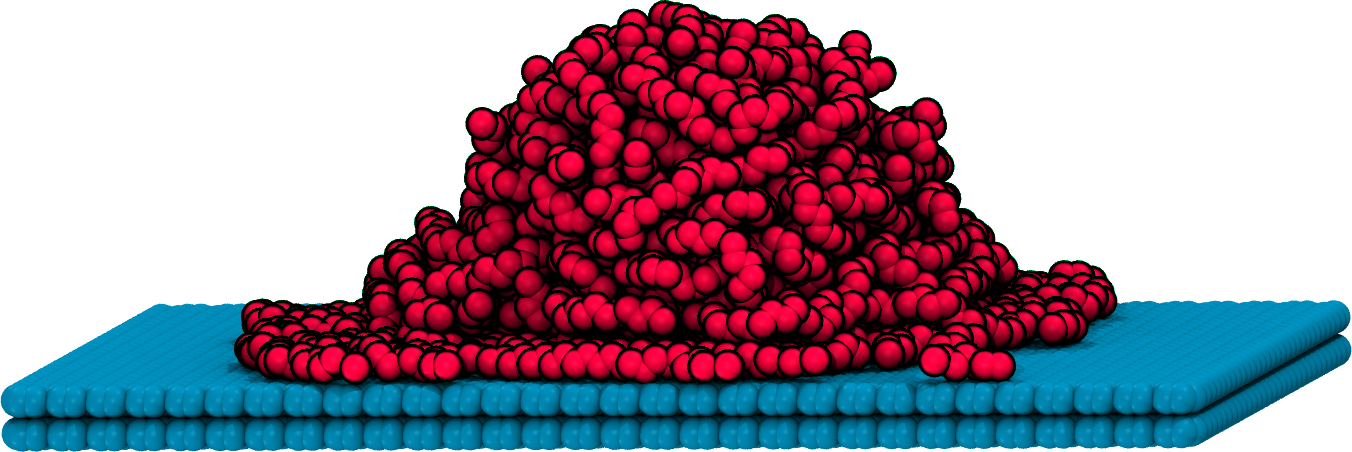
flowerMD's design allows for creation of modules where an end-to-end interface is designed with a complex task in mind, such as fusion welding of polymers and surface wetting:
Installing flowermd from the conda-forge channel can be achieved by adding conda-forge to your channels with:
conda config --add channels conda-forge
conda config --set channel_priority strict
Once the conda-forge channel has been enabled, flowermd can be installed with conda:
conda install flowermd
or with mamba:
mamba install flowermd
Installing from source for development:
Clone this repository:
git clone git@github.com:cmelab/flowerMD.git
cd flowerMD
Set up and activate environment:
conda env create -f environment-dev.yml
conda activate flowermd-dev
python -m pip install -e .
A note on GPU compatibility:
To install a GPU compatible version of HOOMD-blue in your flowerMD environment, you need to manually set the CUDA version before installing flowermd. This is to ensure that the HOOMD build pulled from conda-forge is compatible with your CUDA version. To set the CUDA version, run the following command before installing flowermd:
export CONDA_OVERRIDE_CUDA="[YOUR_CUDA_VERSION]"
Please check out the tutorials for a detailed description of how to use flowerMD and what functionalities it provides.
Documentation is available at https://flowermd.readthedocs.io
If you use flowerMD in your research, please cite the following paper:
Albooyeh, M., Jones, C., Barrett, R., & Jankowski, E. (2023). FlowerMD: Flexible Library of Organic Workflows and Extensible Recipes for Molecular Dynamics. Journal of Open Source Software, 8(92), 5989, https://doi.org/10.21105/joss.05989
We welcome all contributions to flowerMD. Please see contributing guidelines for more information.