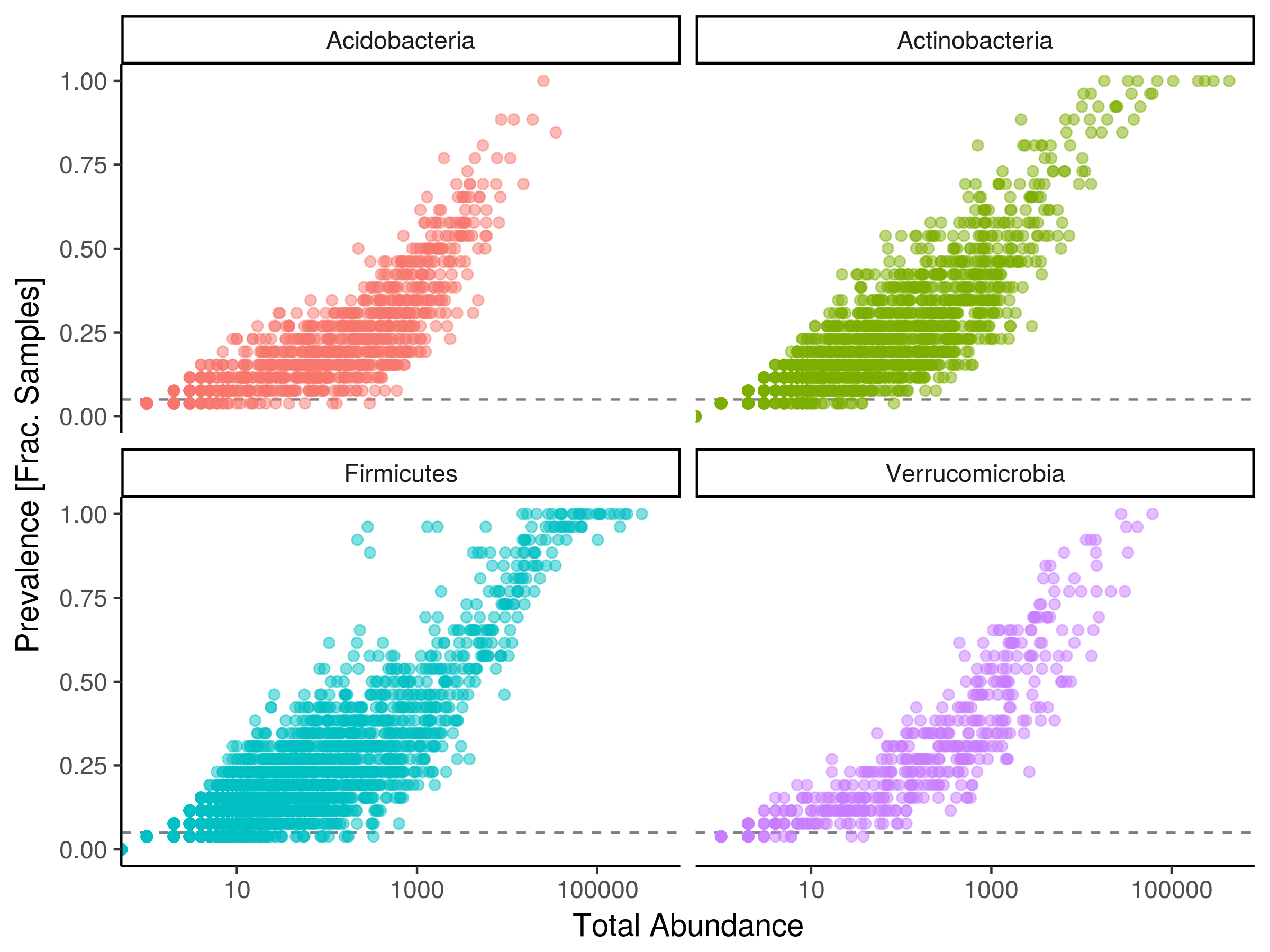
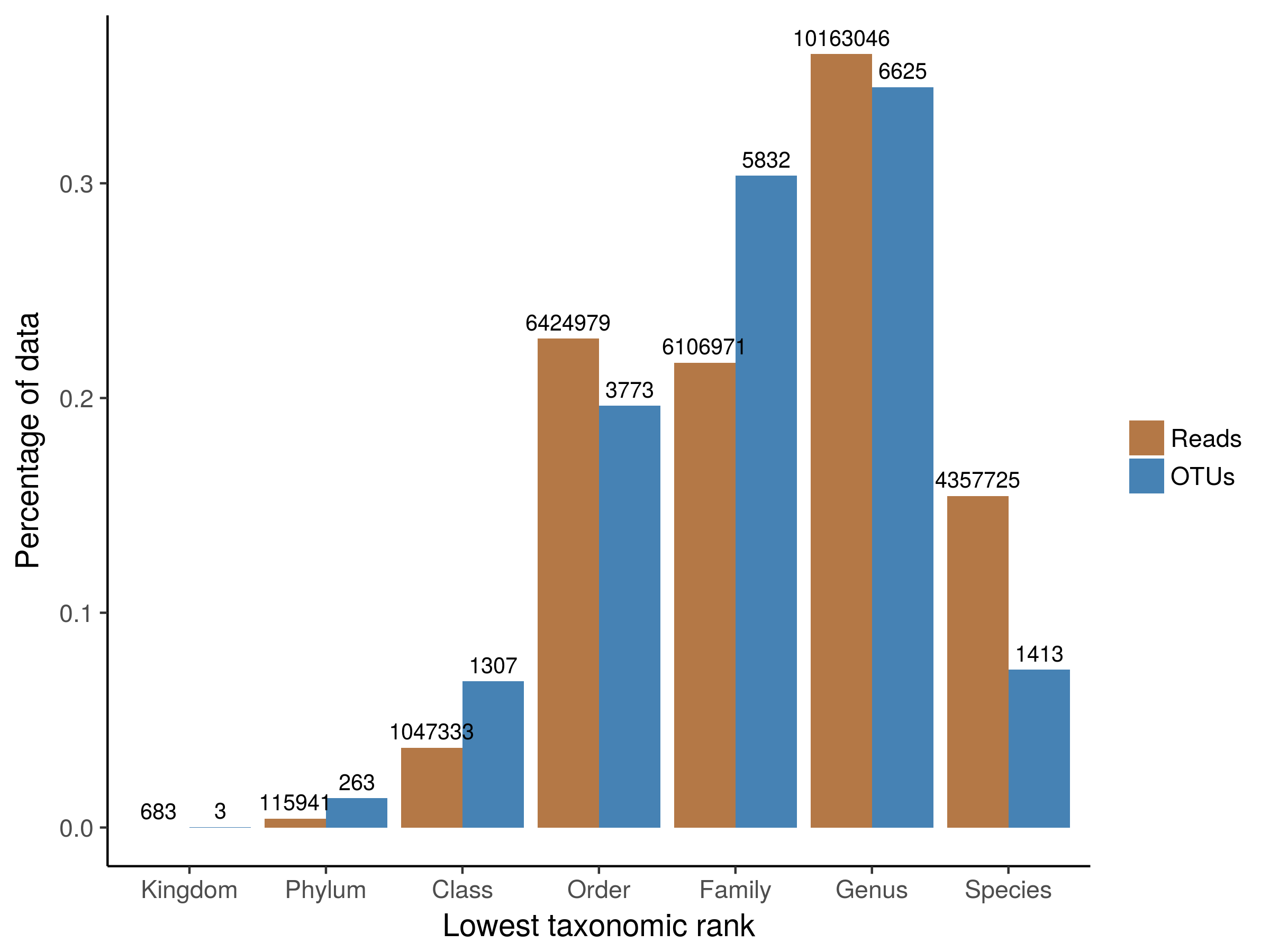
Miscellaneous functions for metagenomic analysis.
The repository is currently in ALPHA state. Nothing is guaranteed and the material is subject to change without a notice (e.g., function names or arguments).
Vignette is under construction.
- Multiple rarefaction
- OTU abundance averaging following CoDa (Compositional Data Analysis) workflow
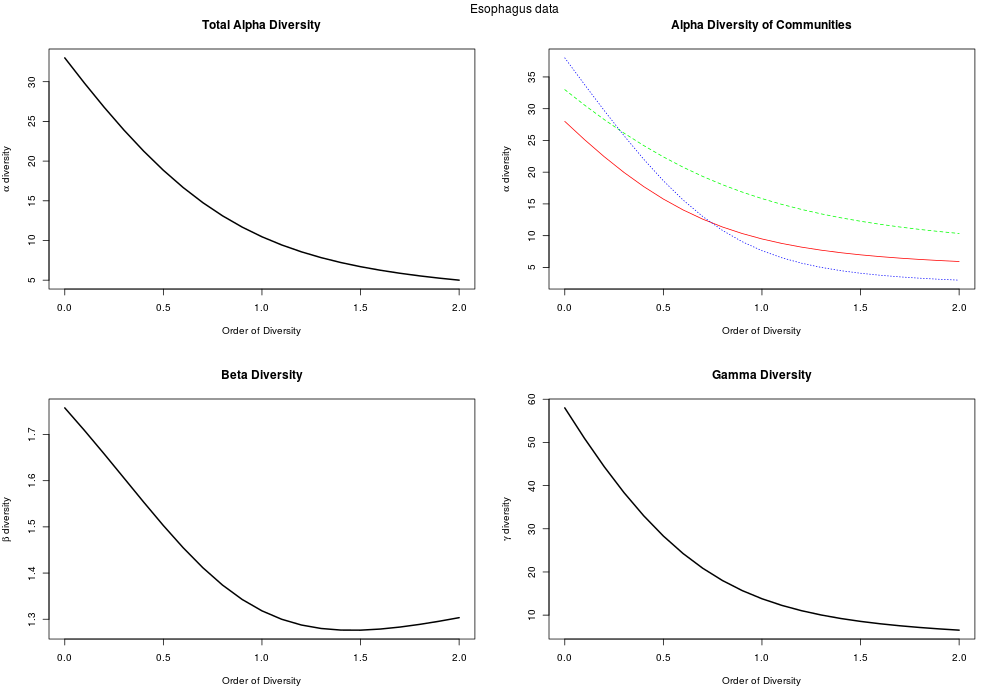
- Phylogenetic diversity estimation (including standardized effect sizes)
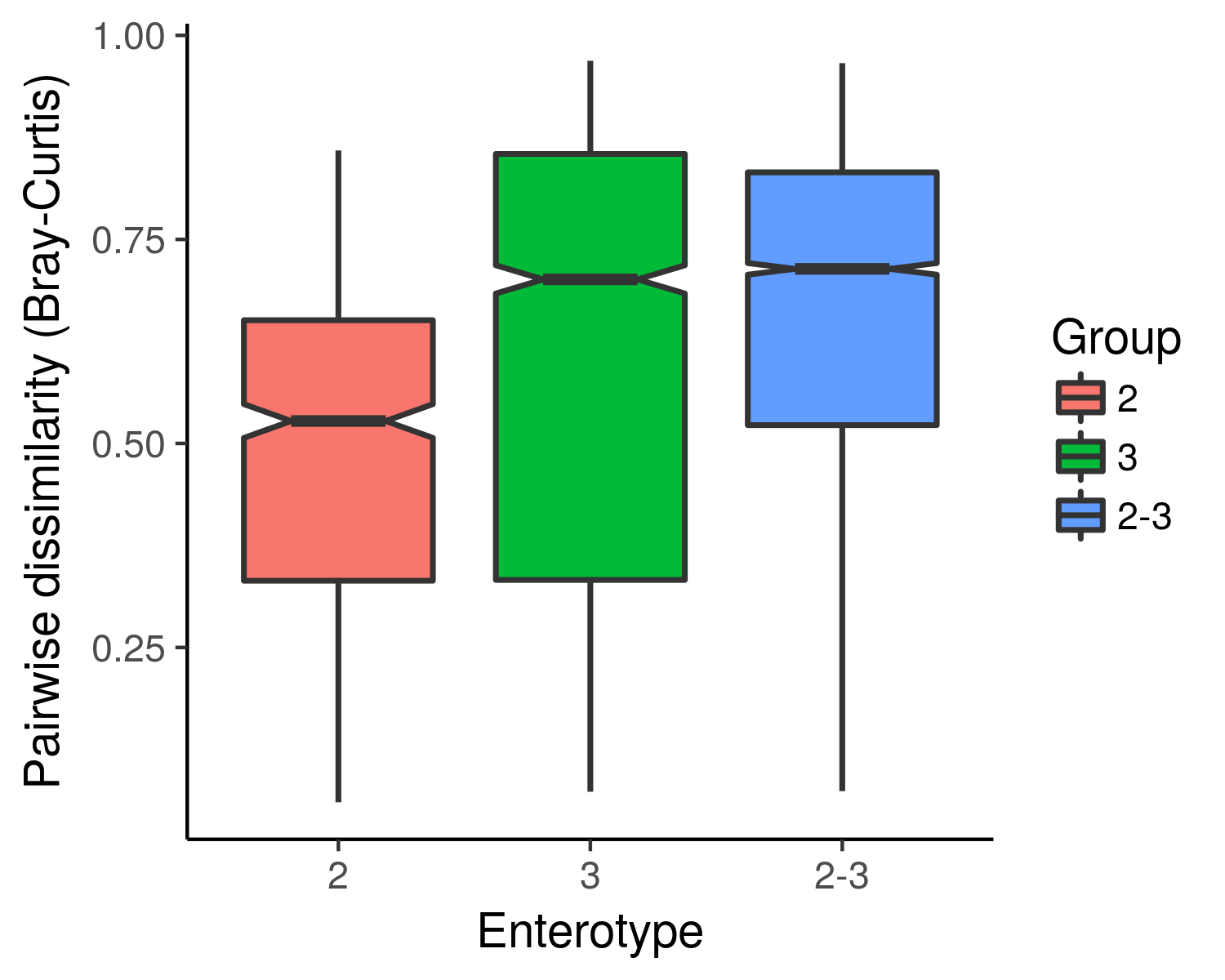
- Pairwise dissimilarity boxplots
devtools::install_github("vmikk/metagMisc")
source("http://bioconductor.org/biocLite.R")
- phyloseq:
biocLite("phyloseq") - dada2:
biocLite("dada2") - ALDEx2:
biocLite("ALDEx2") - metagenomeSeq:
biocLite("metagenomeSeq") - DESeq2:
biocLite("DESeq2") - vegan:
install.packages("vegan") - ggplot2
- plyr
- openssl
metagMisc stands on the shoulders of numerous R-packages (see Dependencies). In particular, it would not have happened without phyloseq and vegan packages. Please cite R and R packages when you use them for data analysis.
The development of this software was supported by RFBR grants 16-04-01259 and 15-29-02765.