Work on this project is ongoing and is described in detail in the original paper.
If you use this work, please cite the following paper:
Meltzer, P., Mallea, M. D. G., & Bentley, P. J. (2019). PiNet: Attention Pooling for Graph Classification, NeurIPS 2019 Graph Representation Learning Workshop
If this work is of interest to you, or you use the code, I'd love your feedback. Please email me at p.meltzer@cs.ucl.ac.uk with any comments, criticisms or suggestions! :D
A Pytorch implementation of Pinet can be found here.
out_dim_a2: output dimension for attentionout_dim_x2: output dimension for featureslearn_pqr: learn message passing parametrisation during training (default isTrue)preprocess_A: List of options as Strings (for manual pre-processing - should not be used withlearn_pqr=True)'add_self_loops''sym_normalise_A''laplacian''sym_norm_laplacian'- could include multiple, i.e.:
['add_self_loops', 'sym_normalise_A']
tensor_board_logging: enable logging for TensorBoardreduce_lr_callback: reduce learning rate based on validation set
from model.PiNet import PiNet
from analysis.experiment2 import generate
from sklearn.model_selection import StratifiedKFold
num_classes = 3
batch_size = 5
pinet = PiNet()
folds = list(StratifiedKFold(n_splits=10, shuffle=True).split(X, Y))A: List of adjacency matrices as ndarraysX: List of features matrices as ndarraysY: (n x 1) ndarray containing class no.
num_classes: number of classesepochs: default 200batch_size: default 50folds: Folds or splits of train/test idsdataset_name: default is 'dataset_name'verbose: default is 1
accs, times = pinet.fit_eval(A, X, Y, num_classes=num_classes,
epochs=50, batch_size=batch_size, folds=folds, verbose=0)
preds = pinet.get_predictions(A, X, Y, batch_size=batch_size)- GCN: model/MyGCN.py
MyGCNlayer takes a list ofA_andX^{l}as input, and gives a single output ofX^{l+1}
params:
num_nodes_per_graph = 50
num_graph_classes = 5
num_node_classes = 2
num_graphs_per_class = 100
batch_size = 5
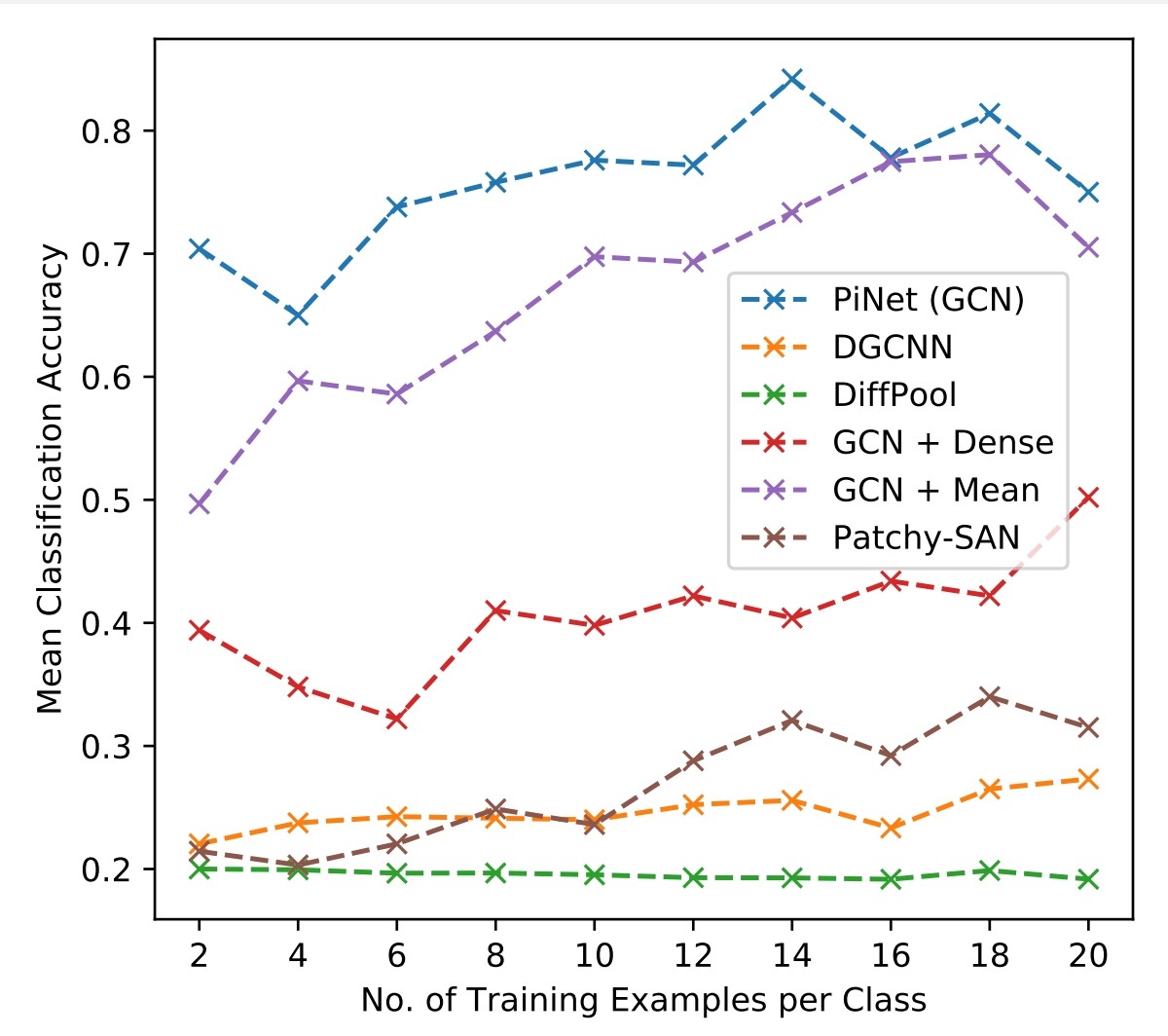
examples_per_classes = [2, 4, 6, 8, 10]- train set selected by stratified sample
- repeated 10x per
examples_per_classes
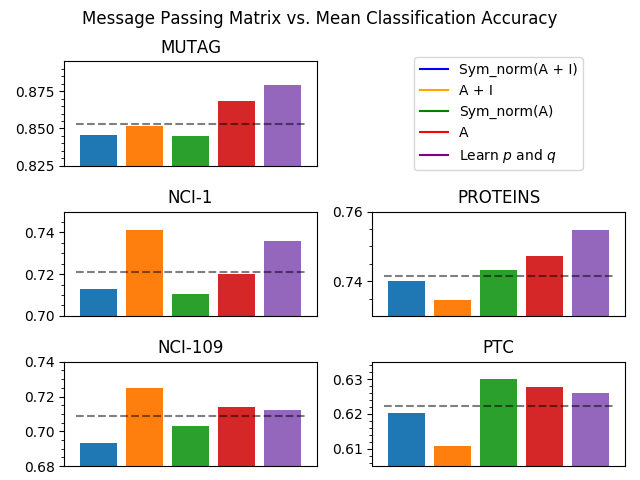
Observe effect of various matrices for message passing/diffusion.
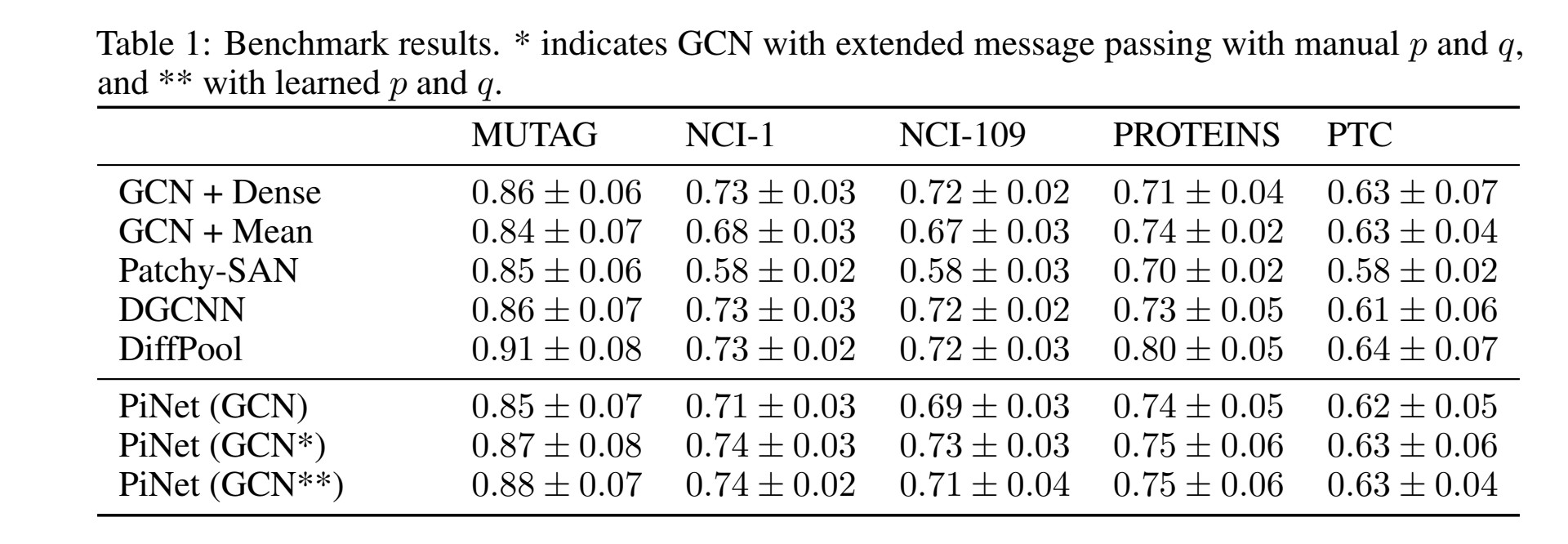
Compare against existing methods on benchmark data.
Mean classification accuracies for each classifier. For manual search the values p and q as follows: MUTAG and PROTEINS p = 1, q = 0, NCI-1 and NCI-109 p = q = 1, PTC p = q = 0. * indicates PiNet (both models) achieved statistically significant gain.