Three commands to start analysing your metagenome data:
conda install -y -c bioconda -c conda-forge metagenome-atlas
atlas init --db-dir databases path/to/fastq/files
atlas run all
All databases and dependencies are installed on the fly in the directory databases.
You want to run these three commands on the example data. If you have more time, then we recommend you configure atlas according to your needs.
- check the
samples.tsv - edit the
config.yaml - run atlas on any cluster system For more details see documentation.
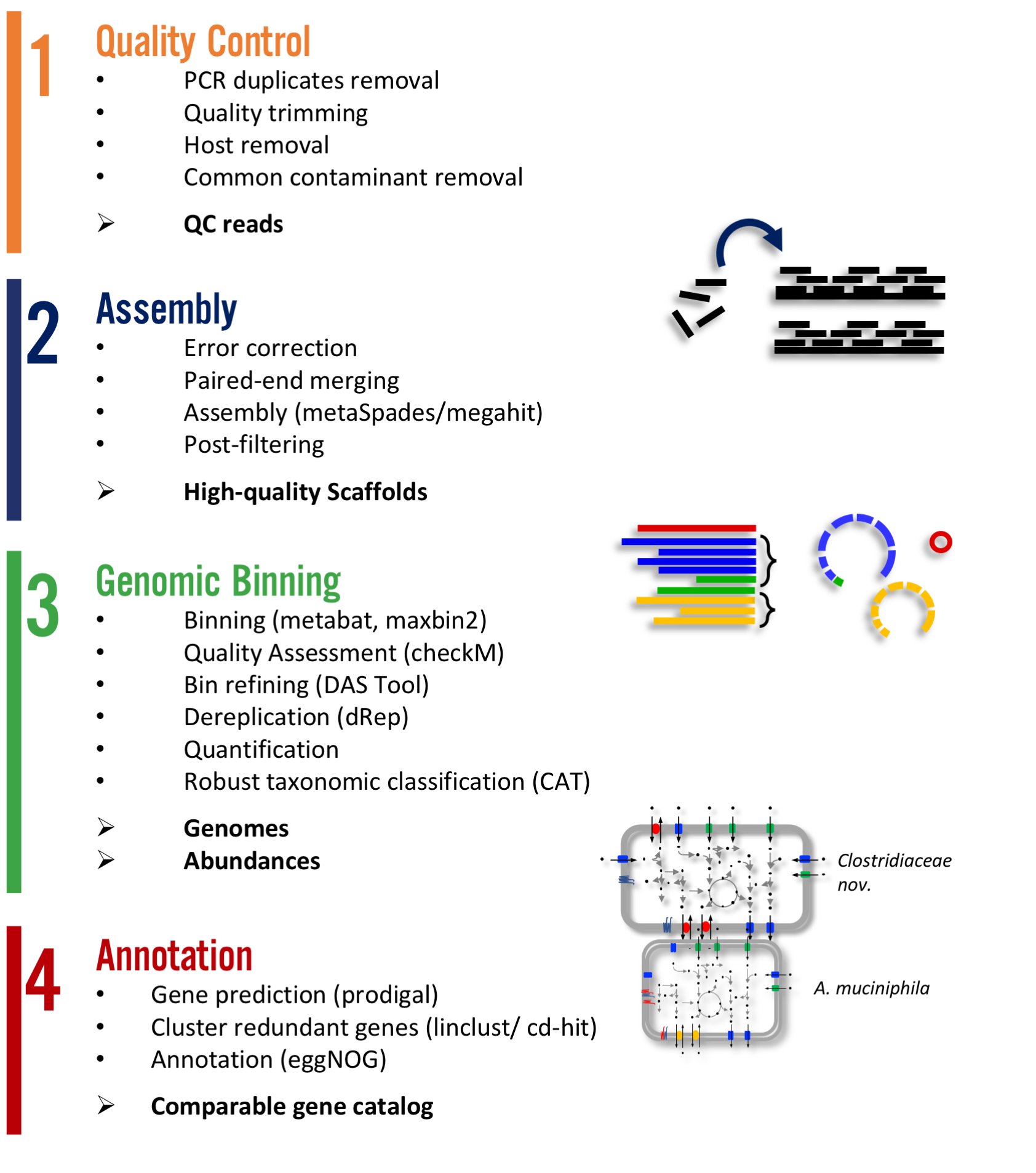
Atlas is a easy to use metagenomic pipeline
Atlas should be run on a linux sytem, with enough memory (min ~50GB but assembly usually requires 250GB). The only dependency is the conda package manager, which can easy be installed with anaconda. We recommend you to create a conda environment for atlas to avoid any conflicts of versions.
conda create -y -n atlasenv
source activate atlasenv
conda install -y -c bioconda -c conda-forge metagenome-atlas
And you can run atlas. All other dependencies are installed in specific environments during the run of the pipeline.
For local execution we have also a docker container
BSD-3.
ATLAS: a Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data.
Kieser, S., Brown, J., Zdobnov, E. M., Trajkovski, M. & McCue, L. A.
BMC Bioinformatics 21, 257 (2020).
doi: 10.1186/s12859-020-03585-4