Efficient and accurate pathogenicity prediction for coding and regulatory structural variants in long-read genome sequencing.
Most users should download the latest SvAnna distribution ZIP file from the Releases page.
SvAnna is a standalone command-line Java application and can be run as follows:
java -jar svanna-cli.jar -d path/to/svanna/data \
-t HP:0008330 \
--vcf example.vcf.gz \
--output-format html,csv,vcfThe analysis will filter out common SVs and perform phenotype-driven prioritization of the remaining SVs. The SVs are assigned with "Pathogenicity of Structural variation" (PSV) score and written into one of several output formats, such as CSV table, a VCF file, or a detailed HTML report.
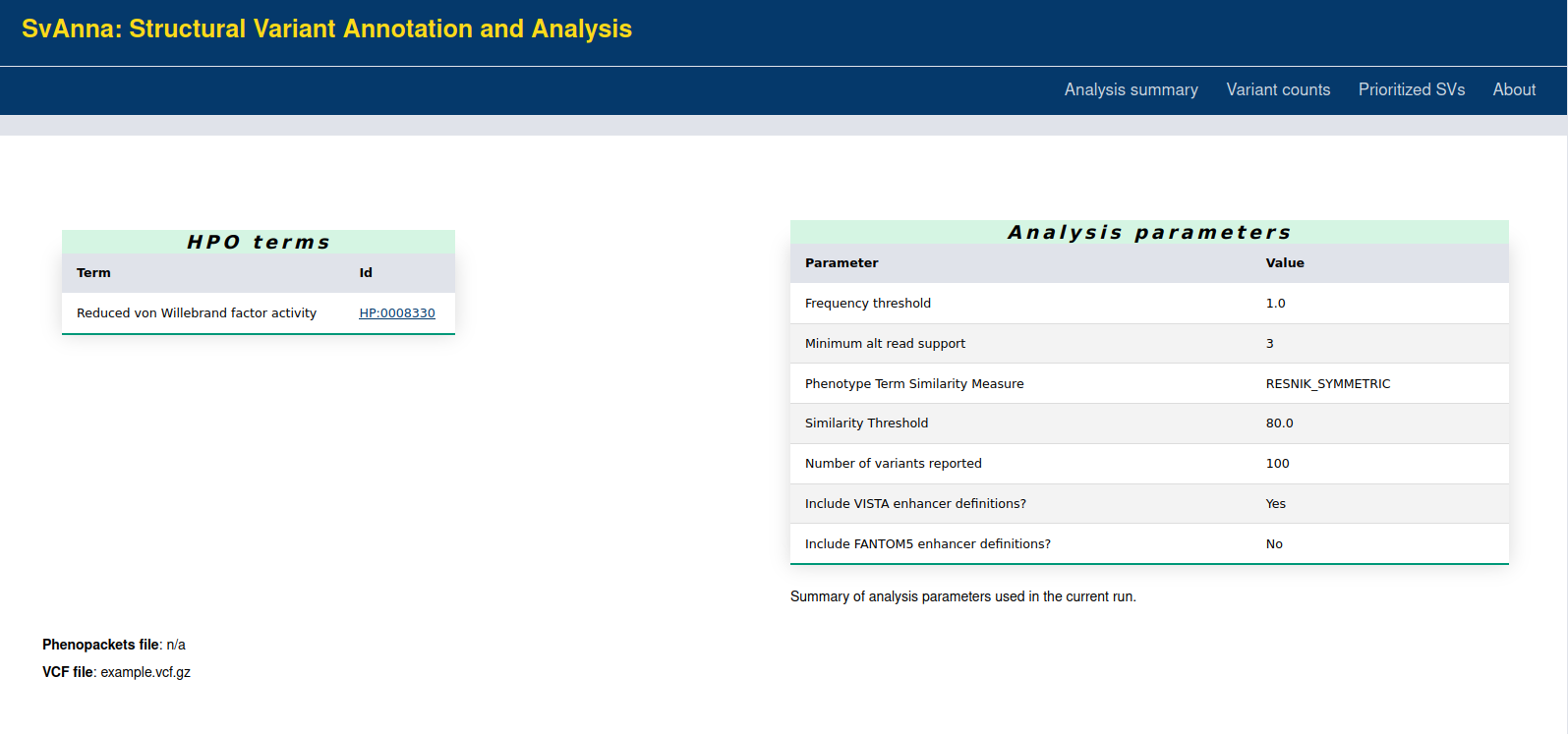
The HTML report includes a header with the analysis summary and the SVs ordered by the PSV score with the best scores on top.
The summary presents the clinical features encoded into terms of Human Phenotype Ontology (HPO) as well as the other analysis parameters.
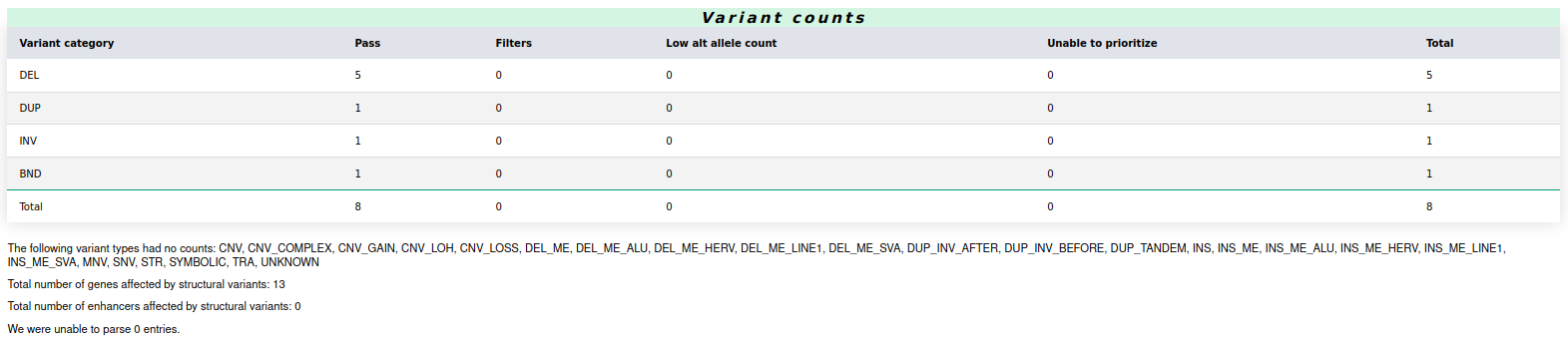
The report further breaks down SVs into several categories:
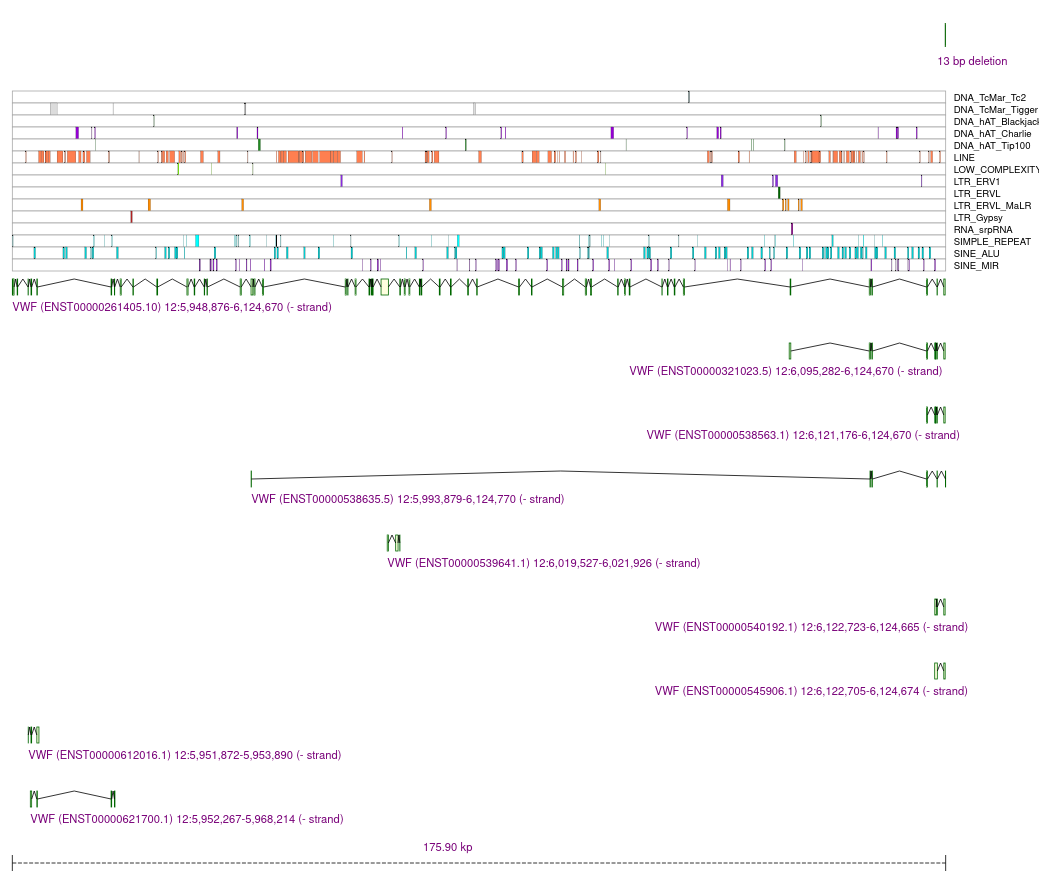
Last, each SV is presented in the context of the overlapping genes and transcripts:

We also show the variant in context of the neighboring repetitive regions and genes/transcripts:
Please consult the Read the docs site for a detailed documentation:
- stable version describing the latest release at the Releases page, or
- latest version summarizing the latest development on
developmentbranch.
Check out SvAnna manuscript in Genome Medicine.