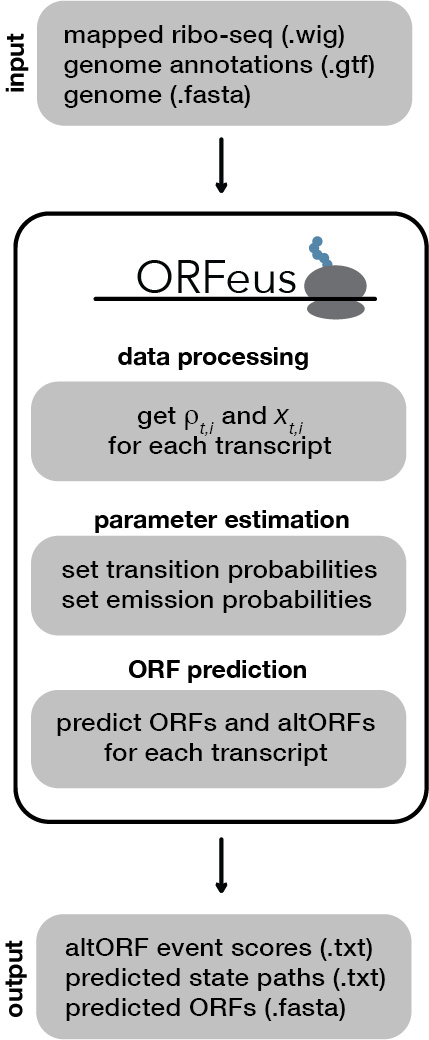
ORFeus is an ORF prediction tool designed to detect alternative translation events, including programmed ribosomal frameshifts, stop codon readthrough, and short upstream or downstream ORFs. It requires aligned ribosome profiling (ribo-seq) reads, reference transcript annotations, and a reference genome. ORFeus can be run on both bacterial and eukaryotic data.
Note that high-resolution (even nucleotide-resolution) ribosome profiling data is ideal. The higher-resolution the data and the deeper the sequencing, the better predictions ORFeus will make.
Build the model:
python build.py forward.wig reverse.wig genome.fa annotations.gtf
Run the model:
python run.py data.txt.gz parameters_h1.npy parameters_h0.npy
ORFeus requires the following input files. (See input files for details on preparing these to work with ORFeus, particularly the aligned ribo-seq reads file.)
- Aligned ribo-seq read counts (format: wiggle or bedgraph)
- Transcript annotations (format: gtf or gff)
- Genome (format: fasta)
- You are interested in finding alternative translation events in an annotated species. (Run ORFeus and look at the top predictions.)
- You want to find changes in translation across multiple conditions or timepoints. (Run ORFeus separately on ribo-seq data from each condition and compare predictions.)
- You need a de novo ORF caller for a novel genome. (ORFeus requires input transcript annotations to determine transcript bounds and for training parameters.)
- You don't have high-resolution ribo-seq data. (ORFeus infers translation based on ribosome profiling reads.)
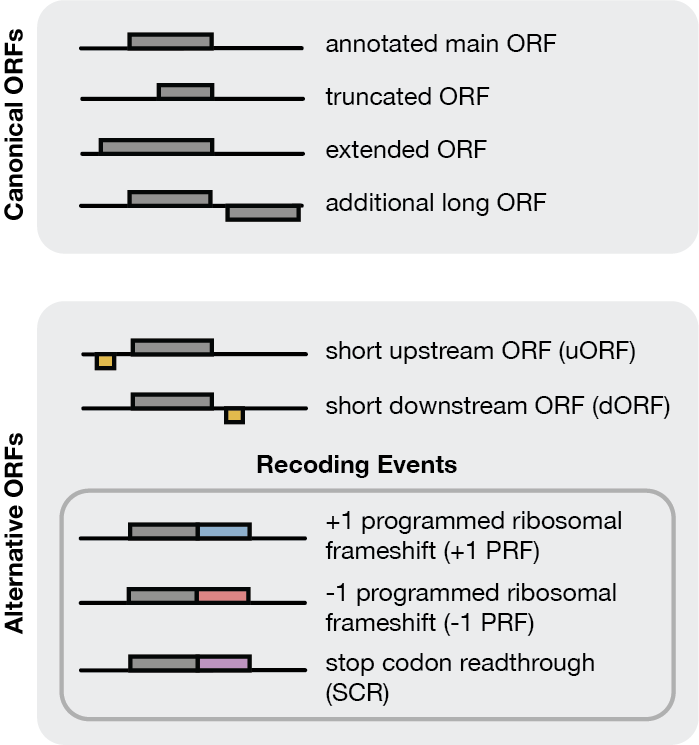
ORFeus can predict the following types of canonical and alternative ORFs:
We recommend installing Anaconda, which is a Python distribution that comes with all of these packages.
git clone https://github.com/morichardson/ORFeus
Navigate to the ORFeus directory:
cd ORFeus
Create and activate an environment with python 3.7 and the required versions of python packages (suggested):
conda create --name orfeus_env --file requirements.txt
conda activate orfeus_env
Set the ORFeus parameters:
python orfeus/build.py data/coronavirus/Finkel2021_forward.wig data/coronavirus/Finkel2021_reverse.wig data/coronavirus/dna.fa data/coronavirus/annotations.gtf -o data/coronavirus/
Run ORFeus:
python orfeus/run.py data/coronavirus/data.txt.gz data/coronavirus/parameters_h1.npy data/coronavirus/parameters_h0.npy -o data/coronavirus/
When finished running ORFeus, you can deactivate the python environment (suggested):
conda deactivate
The annotations file must be in GTF or GFF
format.
At minimum, the annotations file must contain the following columns
(placeholder columns are not used by ORFeus and can be populated with any value,
though the standard is .):
seqname: name of the chromosome (note this must match exactly theseqnamein the genome sequence file)source: placeholderfeature: feature type (note onlyfive_prime_utr,exon, andthree_prime_utrfeatures will be kept)start: first position of the feature, 1-indexedend: last position of the feature, 1-indexedscore: placeholderstrand: + (forward) or - (reverse)frame: placeholderattribute: semicolon-separated list with additional information (note only the info below will be kept)transcript_idtranscript_nametranscript_biotype(note only"protein_coding"features will be kept)
Please note that any chromosomes (seqname) column included in the annotations
file will be considered in by ORFeus when setting parameters. Be sure to remove
any extra chromosomes or scaffolds you don't want considered in the final
analysis (e.g. mitochondrial chromosomes should be removed if your ribo-seq
data is for cytosolic RNA) This is very important to make sure the parameters
are set correctly. Otherwise added noise from these extra chromosomes is
incorporated into the model.
Below is an example transcript from a GTF file that meets the minimum requirements. All placeholder fields have been populated with a period.
V . five_prime_utr 546794 546816 . + . transcript_id "YER178W_mRNA"; transcript_name "PDA1"; transcript_biotype "protein_coding";
V . exon 546817 548079 . + . transcript_id "YER178W_mRNA"; transcript_name "PDA1"; transcript_biotype "protein_coding";
V . three_prime_utr 548080 548208 . + . transcript_id "YER178W_mRNA"; transcript_name "PDA1"; transcript_biotype "protein_coding";
The genome sequence file must be in
FASTA format. There should
be one sequence entry for each unique seqname (chromosome) in the annotations
file.
Below is an example FASTA file excerpt for the chromosome of the above
example transcript. Note that the seqname matches the seqname column entries
in the annotations example.
>V dna:chromosome chromosome:R64-1-1:V:1:576874:1 REF
CGTCTCCTCCAAGCCCTGTTGTCTCTTACCCGGATGTTCAACCAAAAGCTACTTACTACC
TTTATTTTATGTTTACTTTTTATAGATTGTCTTTTTATCCTACTCTTTCCCACTTGTCTC
TCGCTACTGCCGTGCAACAAACACTAAATCAAAACAGTGAAATACTACTACATCAAAACG
CATATTCCCTAGAAAAAAAAATTTCTTACAATATACTATACTACACAATACATAATCACT
...
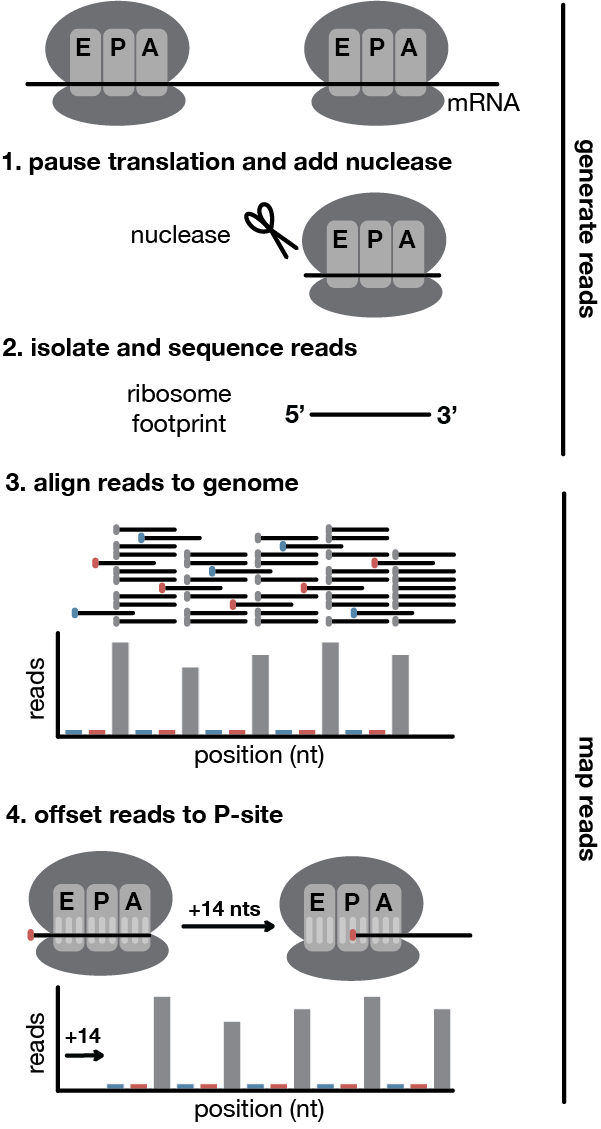
The final aligned ribo-seq read counts must be in either WIG format or BedGraph format. The raw reads must be aligned and then the read ends should be offset to correspond to the P-site of the ribosome. The count of read ends at each position of the genome should be stored, with one file for the forward strand and one for the reverse strand.
Align raw ribo-seq reads to the genome. Filter out reads mapping to annotated ncRNA sequences. You should decide whether uniqely-mapping or multi-mapping is appropriate for your data set.
Uniquely-mapping reads:
- filters out reads that map to repetitive regions (e.g. regions with repeated sequences may appear as gaps in the read density, even though they may actually be translated)
- filters out reads that map to similar or related sequences (e.g. insertion sequences that have multiple copies in the genome will have no reads, even though they may actually be translated)
Multi-mapping reads:
- generates confounding signals from mis-mapped multi-mapping reads (e.g. reads that were actually generated from one transcript also map to another transcript, adding noise)
- complicates interpretation of predictions (e.g. predictions of alternative events may be due to reads from that transcript or another transcript)
In some cases, you may want to run ORFeus twice: once on the uniquely-mapped reads and once on the multi-mapped reads. This will allow you to compare the predictions and determine which events may be artifacts of read mapping. Any predictions that differ between the two runs should be examined more closely, since they might arise from mapping artifacts.
Before passing the data to ORFeus, you need to offset the read ends so they align to a position within the P-site of the ribosome. This lets ORFeus infer the exact codon being translated for each read.
You can determine the offset for each read length and export the resulting read counts using existing software packages like Shoelaces or using your own custom scripts.
python orfeus/build.py forward.wig reverse.wig genome.fa annotations.gtf
Building the model involves processing the data and setting the model parameters
(transition probabilities, emission probabilities).
Run python orfeus/build.py -h to see all available options.
usage: build.py [-h] [-s SKIP [SKIP ...]] [--data_file DATA_FILE] [-o OUTDIR]
[-a ALPHA] [-b BETA] [-g GAMMA] [-d DELTA] [-z ZETA] [--f5 F5]
[--f3 F3] [--utr5 UTR5] [--utr3 UTR3] [--orf ORF]
[--uorf UORF] [--dorf DORF]
[--start_codons [START_CODONS [START_CODONS ...]]]
[--sense_codons [SENSE_CODONS [SENSE_CODONS ...]]]
[--stop_codons [STOP_CODONS [STOP_CODONS ...]]] [-r BINS] [-l]
[-m MIN] [-M MAX] [-p] [-f] [-c] [--iters ITERS]
[--window WINDOW] [--threads THREADS]
plus_file minus_file seqs_file annotations_file
Build ORFeus model
positional arguments:
plus_file riboseq plus strand bg/wig file path
minus_file riboseq minus strand bg/wig file path
seqs_file genome or transcriptome fasta file path
annotations_file annotations gtf/gff file path
optional arguments:
-h, --help show this help message and exit
-s SKIP [SKIP ...], --skip SKIP [SKIP ...]
list known frameshifted genes that should be filtered
out before codon assignments
--data_file DATA_FILE
processed data file path
-o OUTDIR, --outdir OUTDIR
output directory for all intermediate and results
files
-a ALPHA, --alpha ALPHA
probability of PRF
-b BETA, --beta BETA probability of -1 PRF given PRF
-g GAMMA, --gamma GAMMA
probability of stop codon readthrough
-d DELTA, --delta DELTA
probability of upstream or downstream short ORFs
-z ZETA, --zeta ZETA probability of multiple non-overlapping ORFs
--f5 F5 fraction of transcripts with 5'UTRs (default:
calculated from annotations)
--f3 F3 fraction of transcripts with 3'UTRs (default:
calculated from annotations)
--utr5 UTR5 mean nucleotide length of 5'UTRs (default: calculated
from annotations)
--utr3 UTR3 mean nucleotide length of 3'UTRs (default: calculated
from annotations)
--orf ORF mean nucleotide length of main ORFs (default:
calculated from annotations)
--uorf UORF mean nucleotide length of upstream short ORFs
(suggested: 50)
--dorf DORF mean nucleotide length of downstream short ORFs
(suggested: 50)
--start_codons [START_CODONS [START_CODONS ...]]
list of valid start codons (default: infer from the
annotations)
--sense_codons [SENSE_CODONS [SENSE_CODONS ...]]
list of valid sense codons (default: all codons except
TAA, TAG, TGA)
--stop_codons [STOP_CODONS [STOP_CODONS ...]]
list of valid stop codons (default: TAA, TAG, TGA)
-r BINS, --bins BINS number of bins to group emission probabilities
(suggested: 25)
-l, --log whether to bin the riboseq values in logspace
(suggested: false)
-m MIN, --min MIN minimum reads per transcript (default: -1)
-M MAX, --max MAX maximum reads per transcript (default: infinity)
-p, --pool whether to pool the observed riboseq values from all
codons of a given state type (suggested: false, unless
you have a small transcriptome with <=20 ORFs)
-f, --fit whether to fit a log-normal distribution to the
observed riboseq values or use the raw frequencies
(suggested: false, unless you have a small
transcriptome with <=20 ORFs)
-c, --coverage whether to run the coverage threshold simulation
--iters ITERS number of iterations to simulate
--window WINDOW window (in nts) around true event to count as correct
altORF prediction
--threads THREADS number of processes to run in parallel when simulating
transcripts
python orfeus/run.py data.txt.gz parameters_h1.npy parameters_h0.npy
Running the model generates the predicted ORFs and altORFs, using the given
processed data and model parameters (generated by orfeus/build.py)
The most likely transcript path is calculated using the Viterbi
algorithm. Run python orfeus/run.py -h to see all available options.
usage: run.py [-h] [-o OUTDIR] [-c COVERAGE] [--threads THREADS]
data_file parameters_file_h1 parameters_file_h0
Run ORFeus model
positional arguments:
data_file processed data file path (generated by
orfeus_build_model.py)
parameters_file_h1 model parameters file path (generated by
orfeus_build_model.py)
parameters_file_h0 null model parameters file path (generated by
orfeus_build_model.py)
optional arguments:
-h, --help show this help message and exit
-o OUTDIR, --outdir OUTDIR
output directory for all intermediate and results
files
-c COVERAGE, --coverage COVERAGE
mean riboseq coverage threshold
--threads THREADS number of processes to run in parallel when running
the model on all transcripts
A table of predicted ORFs and altORFs is generated in results.txt. This table
contains the following columns for each transcript:
transcript_id: transcript IDtranscript_name: transcript name (Noneif missing)mean_coverage: mean number of ribo-seq reads per nucleotide for the transcriptpredicted=annotated:trueif the predicted ORFs are exactly the same as the annotated ORFs,falseotherwiseannotated_events: annotated ORF and altORF events (indices for each event in parentheses, 1-indexed and inclusive of start and end index)predicted_events: predicted ORF and altORF events (indices for each event in parentheses, 1-indexed and inclusive of start and end index)log_odds_score: log of the predicted path score divided by the null path score (see below)annotated_path_score: total probability of the annotated pathpredicted_path_score: total probability of the predicted path (path if altORFs events are allowed)null_path_score: total probability of the best canonical path (path if no altORF events are allowed)
Below is an example of the predicted ORF and altORF output for three transcripts.
transcript_id transcript_name mean_coverage predicted=annotated annotated_events predicted_events log_odds_score annotated_path_score predicted_path_score null_path_score
YOR239W_mRNA ABP140 2.1462 false +1 PRF (829-829), ORF (1-828), ORF (830-1888) 789.118 nan -3568.647 -4357.765
YEL009C_mRNA GCN4 0.3338 false ORF (572-1417) uORF (211-222), uORF (279-287), uORF (396-407), uORF (421-432), ORF (572-1417) 18.157 -2483.361 -2465.204 -2483.361
YAR028W_mRNA None 1.0389 true ORF (55-759) ORF (55-759) 0.000 -1962.025 -1962.025 -1962.025
The output predicted mRNA sequence file is in FASTA format. The translation of each transcript is denoted for each sequence entry. We use with the following symbols to denote each type of ORF and altORF feature:
.: nucleotide in UTR (this symbol is shown in place of each UTR nucleotide)[X: beginning of a translated ORF (this symbol appears before the first nucleotide of the start codon X)X]: end of a translated ORF (this symbol appears after the last nucleotide of the stop codon X)X|Y: stop codon readthrough (this symbol appears after between the last nucleotide of the stop codon X and the first nucleotide of the downstream sequence Y)(X): nucleotide skipped during a +1 programmed ribosomal frameshift (these symbols surround nucleotide X that is skipped)(X)(Y): nucleotides skipped during a +2 frameshift, which is how a -1 programmed ribosomal frameshift is represented by the model (these symbols surround nucleotides X and Y that are skipped)
Below is an example FASTA file excerpt for a predicted mRNA ORF sequence.
>YEL009C_mRNA (GCN4) orfeus prediction
............................................................
............................................................
............................................................
..............................[ATGGCTTGCTAA]................
........................................[ATGTGTTAA].........
............................................................
.......................................[ATGTACCCGTAG].......
......[ATGTTTCCGTAA]........................................
............................................................
.......................................[ATGTCCGAATATCAGCCAAG
TTTATTTGCTTTAAATCCAATGGGTTTCTCACCATTGGATGGTTCTAAATCAACCAACGA
AAATGTATCTGCTTCCACTTCTACTGCCAAACCAATGGTTGGCCAATTGATTTTTGATAA
ATTCATCAAGACTGAAGAGGATCCAATTATCAAACAGGATACCCCTTCGAACCTTGATTT
TGATTTTGCTCTTCCACAAACGGCAACTGCACCTGATGCCAAGACCGTTTTGCCAATTCC
GGAGCTAGATGACGCTGTAGTGGAATCTTTCTTTTCGTCAAGCACTGATTCAACTCCAAT
GTTTGAGTATGAAAACCTAGAAGACAACTCTAAAGAATGGACATCCTTGTTTGACAATGA
CATTCCAGTTACCACTGACGATGTTTCATTGGCTGATAAGGCAATTGAATCCACTGAAGA
AGTTTCTCTGGTACCATCCAATCTGGAAGTCTCGACAACTTCATTCTTACCCACTCCTGT
TCTAGAAGATGCTAAACTGACTCAAACAAGAAAGGTTAAGAAACCAAATTCAGTCGTTAA
GAAGTCACATCATGTTGGAAAGGATGACGAATCGAGACTGGATCATCTAGGTGTTGTTGC
TTACAACCGCAAACAGCGTTCGATTCCACTTTCTCCAATTGTGCCCGAATCCAGTGATCC
TGCTGCTCTAAAACGTGCTAGAAACACTGAAGCCGCCAGGCGTTCTCGTGCGAGAAAGTT
GCAAAGAATGAAACAACTTGAAGACAAGGTTGAAGAATTGCTTTCGAAAAATTATCACTT
GGAAAATGAGGTTGCCAGATTAAAGAAATTAGTTGGCGAACGCTGA].............
...........................................................
The output predicted protein sequence file is in FASTA format. The protein translation of each predicted ORF is a separate entry in the file.
Below is an example FASTA file excerpt for the predicted protein sequences for the above example transcript.
>YEL009C_mRNA (GCN4) orfeus prediction 0
MAC
>YEL009C_mRNA (GCN4) orfeus prediction 1
MC
>YEL009C_mRNA (GCN4) orfeus prediction 2
MYP
>YEL009C_mRNA (GCN4) orfeus prediction 3
MFP
>YEL009C_mRNA (GCN4) orfeus prediction 4
MSEYQPSLFALNPMGFSPLDGSKSTNENVSASTSTAKPMVGQLIFDKFIKTEEDPIIKQD
TPSNLDFDFALPQTATAPDAKTVLPIPELDDAVVESFFSSSTDSTPMFEYENLEDNSKEW
TSLFDNDIPVTTDDVSLADKAIESTEEVSLVPSNLEVSTTSFLPTPVLEDAKLTQTRKVK
KPNSVVKKSHHVGKDDESRLDHLGVVAYNRKQRSIPLSPIVPESSDPAALKRARNTEAAR
RSRARKLQRMKQLEDKVEELLSKNYHLENEVARLKKLVGER