Originally published by Soledad Galli @ Train in Data in May, 2021.
Modified by Mikel Sagardia while folowing the associated Udemy course Hyperparameter Optimization for Machine Learning.
-
Cross-Validation
- K-fold, LOOCV, LPOCV, Stratified CV
- Group CV and variants
- CV for time series
- Nested CV
-
Basic Search Algorithms
- Manual Search, Grid Search and Random Search
-
Bayesian Optimization
- with Gaussian Processes
- with Random Forests (SMAC) and GBMs
- with Parzen windows (Tree-structured Parzen Estimators or TPE)
-
Multi-fidelity Optimization
- Successive Halving
- Hyperband
- BOHB
-
Other Search Algorthms
- Simulated Annealing
- Population Based Optimization
-
Gentetic Algorithms
- CMA-ES
-
Python tools
- Scikit-learn
- Scikit-optimize
- Hyperopt
- Optuna
- Keras Tuner
- SMAC
- Others
- Added by me: Ax
- Hyperparameter tuning for Machine Learning
- Overview of Topics
- Table of Contents
- Setup
- Section 2: Hyperparameter Tuning: Overview
- Section 3: Performance Metrics
- Section 4: Cross-Validation
- Section 5: Basic Search Algorithms: Grid and Random
- Section 6: Bayesian Optimization with Scikit-Optimize
- Bayesian Inference
- Bayes Rule
- Sequential Model-Based Optimization (SMBO)
- Literature
- Scikit-Optimize for Bayesian Optimization: Notes
- Example: Manual Gaussian Optimization of a Black Box 1D Function
- Example: Manual Gaussian Optimiation of a Grandient Boosted Tree with 1 Hyperparameter
- Example: Manual Gaussian Optimization of a Grandient Boosted Tree with 4 Hyperparameters
- Example: Automatic Gaussian Optimization of a Grandient Boosted Tree with 4 Hyperparameters (BayesSearchCV)
- Example: Bayes Optimization with Different Kernels (Manual)
- Example: Manual Bayesian Optimization of an XGBoost Classifier
- Example: Manual Bayesian Optimization of a Keras-CNN
- Section 7: Other Sequential Model-Based Optimization (SMBO) Methods
- Section 8: Multi-Fidelity Optimization
- Section 9: Empty
- Section 10: Scikit-Optimize Review and Summary
- Section 11: Hyperopt Review and Summary
- Section 12: Optuna Guide
- Section 13: Ax Platform Guide
Links:
Environment:
conda create --name hyp pip python=3.8
conda activate hyp
python -m pip install -r requirements.txtUsed datasets:
- MNIST Kaggle: CNN for classification.
- Breast Cancer Wisconsin (Diagnostic): linear and non-linear models (trees) for classification.
- Boston housing dataset: regression.
from sklearn.datasets import load_breast_cancer
from sklearn.datasets import load_boston
# MNIST Kaggle dataset
# https://www.kaggle.com/c/digit-recognizer/data
data = pd.read_csv("../data/train.csv")
# Breast cancer dataset
breast_cancer_X, breast_cancer_y = load_breast_cancer(return_X_y=True)
X = pd.DataFrame(breast_cancer_X)
y = pd.Series(breast_cancer_y).map({0:1, 1:0})
# Boston housing dataset
boston_X, boston_y = load_boston(return_X_y=True)
X = pd.DataFrame(boston_X)
y = pd.Series(boston_y)Hyperparameters are those parameters which are not learnt during taining, but chosen by the user. They can be used to:
- Improve convergence
- Improve performance
- Prevent overfitting
- etc.
However, the hyperparameters affect the parameters learnt by the model.
Typical hyperparameters for Random Forests and Gradient Boosted Trees:
- Number of trees
- The depth of the tree
- Learning rate (GBMs)
- The metric of split quality
- The number of features to evaluate at each node
- The minimum number of samples to split the data further
The most important hyperparameters are those that optimize the generalization error, which is not necessarily the loss.
Some hyperparameters do not affect the model performance; we need to try all possible value combinations, but that comes with an inrceased computational cost.
A hyperparameter search consists of:
- Hyperparameter space: the hyperparameters we are going to test and their possible values.
- A method for sampling candidate hyperparameters
- Manual Search
- Grid Search
- Random Search
- Bayesian Optimization
- Other
- A cross-validation scheme
- A performance metric to minimize (or maximize)
Two important concepts that come up often when talking about hyperparameters optimization:
- Hyperparameter response surface: that's the value of the decision metric as function of the hyperparameter values; we want to minimize it. Notation:
lambda = argmin(Phi(lambda)), i.e.,lambdaare the hyperparameter values andPhi(lambda)is the response surface, i.e., the decision metric. - Low effective dimension: the repsonse surface is very sensitive to some hyperparameters and it doesn't change with others; we want to find which hyperparameters affect
Phi.
We can evaluate these two concepts, e.g., when we use the GridSearchCV from Scikit-Learn. These notebooks show how to do that:
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split, GridSearchCV
# model
rf_model = RandomForestRegressor(
n_estimators=100, max_depth=1, random_state=0, n_jobs=4)
# hyperparameter space
rf_param_grid = dict(
n_estimators=[50, 100, 200]
max_depth=[2, 3, 4],
)
# search
# the available metrics can be passed as a string
# https://scikit-learn.org/stable/modules/model_evaluation.html#the-scoring-parameter-defining-model-evaluation-rules
reg = GridSearchCV(rf_model,
rf_param_grid,
scoring='neg_mean_squared_error',
cv=3)
search = reg.fit(X, y)
# best hyperparameters
search.best_params_
# We have all the CV results for each parameter combination
# in this attribute!
search.cv_results_
# mean_fit_time
# std_fit_time
# mean_score_time
# std_score_time
# params
# param_max_depth
# param_n_estimators
# split0_test_score
# split1_test_score
# split2_test_score
# mean_test_score
# std_test_score
# rank_test_score
# Get the CV results
results = pd.DataFrame(search.cv_results_)[['params', 'mean_test_score']]
## 1. Plot response surface
fig = plt.figure(figsize=(8, 8))
ax = plt.axes(projection='3d')
# depth
x = [r['max_depth'] for r in results['params']]
# number of trees
y = [r['n_estimators'] for r in results['params']]
# performance
z = results['mean_test_score']
# plotting
ax.scatter(x, y, z,)
ax.set_title('Response Surface')
ax.set_xlabel('Tree depth')
ax.set_ylabel('Number of trees')
ax.set_zlabel('negative rmse')
plt.show()
## 2. Plot effect
results = pd.DataFrame(search.cv_results_)[['params', 'mean_test_score', 'std_test_score']]
results.index = rf_param_grid['n_estimators']
results['mean_test_score'].plot(yerr=[results['std_test_score'], results['std_test_score']], subplots=True)
plt.ylim(0.2, 0.6)
plt.ylabel('Mean r2 score')
plt.xlabel('Number of trees')
# We can also plot the peformance in the hyperparameter space
results.sort_values(by='mean_test_score', ascending=False, inplace=True)
results.reset_index(drop=True, inplace=True)
results['mean_test_score'].plot(yerr=[results['std_test_score'], results['std_test_score']], subplots=True)
plt.ylim(-0.3, 0)
plt.ylabel('Mean False Negative Rate')
plt.xlabel('Hyperparameter space')Classification metrics:
- Dependent on probability:
- Accuracy: correct / total
- Confusion matrix
- Precision: Positive Predictive Value
- Recall = Sensitivity: True positive rate
- F1
- False Positive Rate
- False Negative Rate
- Accuracy: correct / total
- Independent from probability, aggregate values
- ROC: Receiver-Operator Characteristic Curve
- ROC-AUC: ROC area under the curve
- ROC: Receiver-Operator Characteristic Curve
- Loss:
-(y*log(p) + (1-y)*log(1-p))
Regression metrics:
- Square Error
- Mean Square Error, MSE
- Root Mean Square Error, RMSE
- Mean Absolute Error, MAE
- R2: how much of the total variance that exists in our date is explained by the model
We want to minimize some metrics and maximize others. In Scikit-Learn, the metrics are always maximized; therefore, we use neg_mae, i.e., the -MAE is maximized:
If we want to use a metric which is not defined, i.e., a custom metric defined by us, we can define it with make_scorer. That makes a lot of sense in some cases because there is not an available meric or because we have very specific needs. For instance, in cancer prediction, we want to minimize the false negatives, so we need the False Negative Rate; additionally, we might want to set the probability threshold manually! Choosing the correct metric completely changes both the response surface and the effect of the hyperparameters!
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer, confusion_matrix
def fnr(y_true, y_pred):
"""False negative rate.
This is essential for cases like cancer detection,
in which a false negative has really bad consequences."""
# I we need it, we can pass a probability and define our own threshold:
# y_pred = np.where(y_pred > 0.37, 1, 0)
# BUT we need to set needs_proba=True in make_scorer
tn, fp, fn, tp = confusion_matrix(y_true, y_pred, labels=[0,1]).ravel()
FNR = fn / (tp + fn)
return FNR
fnr_score = make_scorer(
fnr,
greater_is_better=False, # smaller is better
needs_proba=False, # specify y_pred needs to be a probability
)
rf = RandomForestClassifier(n_estimators=100,
max_depth=1,
random_state=0,
n_jobs=4)
# hyperparameter space
params = dict(
n_estimators=[10, 50, 200],
max_depth=[1, 2, 3],
)
# search
# we can use our custom/defined score/metric
clf = GridSearchCV(rf,
params,
scoring=fnr_score,
cv=5)
search = clf.fit(X, y)
# best hyperparameters
search.best_params_There is always a bias-variance tradeoff in our models:
- If the model has bias, it underfits the data, i.e., it is too simplistic and it does not capture essential behaviors.
- If the model has variance, it overfits the data, i.e., it is too complex and it captures noise.
We want a model which generalizes well. To achieve that, we need to evaluate our final model on a test split which has never been seen. When we are choosing between different hyperparameter combinations, we cannot use that test split either, because then we would be fitting the model to that split, i.e., we'd be leaking information from the test set to the training.
To perform a correct hyperparameter tuning, we have two major ways:
- Use 3 splits:
train,validation,test. We train the model withtrainfor each hyperparameter combination and evaluate them withvalidation. Then, we pick the best model and evaluate it withtest. If the model generalizes well, the bestvalidationperformance should be similar to thetestperformance. This approach requires a large dataset; keep in mind that smalltrainsets lead to bad performing models (biased). - We apply cross-validation. We have 2 splits
trainandtest, buttrainis further split inknon-overlapping subsets; then, for each hyperparameter combination, we performktrainings (i.e.,kmodels) in whichk-1subsets are used and the remaining is employed for validation. At the end, we get the mean validation error and thetestsplit is used for the final evaluation.
In gradient descent algorithms, the validation performance can decrease in the beginning but start increasing past a point, in which we start overfitting.
See: Scikit-Learn Cross-Validation
The main goal or strategy is:
- Split the dataset into
trainandtest. - Perform hyperparameter tuning with cross-validation using
train. - Obtain the final performance metrics (with std. dev./error) for
trainandvaland check that they considerably overlap. The validation error/metric is the first estimation of the generalization error. - Compute the performance metrics for the unseen
test; this is the real generalization error. - Verify that the performance of
testis in the confidence interval oftrain.
If the data is independent and identically distributed (iid.), we can use these cross-validation schemes:
- K-fold: the scheme explained before, typically with
k in [5,10]; the larger thek, the more risk of high variance. But try to use at leastk = 5. - Leave-one-out:
k = n, i.e., we leave one data point out for validation. We havenmodels to train, very expensive; additionally, all models have a very similartrainset, which can lead to high variance. It could be used for continuous metrics, but doesn't work well for classification metrics, in general. - Leave-P-out: we leave P points out and carry out combinatorics of points; again, many models, much moe than only leaving one out.
- Repeated K-fold: repeat K-fold
ntimes, each time with data points shuffled. A good alternative. - Stratified K-fold: K-fold for classification in which class ratios are preserved in each folded split. This is important for imbalanced classification problems; try to use it in any classification problem.
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import (
KFold,
RepeatedKFold,
LeaveOneOut,
LeavePOut,
StratifiedKFold,
cross_validate,
train_test_split,
)
# Logistic Regression
logit = LogisticRegression(
penalty ='l2', C=10, solver='liblinear', random_state=4, max_iter=10000)
# K-Fold Cross-Validation
cv = KFold(n_splits=5, shuffle=True, random_state=4)
# Repeated K-Fold Cross-Validation
cv = RepeatedKFold(
n_splits=5,
n_repeats=10,
random_state=4,
)
# Leave One Out Cross-Validation
cv = LeaveOneOut()
# Leave P Out Cross-Validation
cv = LeavePOut(p=2)
# Leave P Out Cross-Validation
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=4)
# estimate generalization error
clf = cross_validate(
logit,
X_train,
y_train,
scoring='accuracy',
return_train_score=True,
cv=cv, # we could also pass an int, and it performs k-fold with n_splits
)
# the score in the subset that was left; k values
clf['test_score']
# the score of the subset used for training; k values
clf['train_score']
# The idea is that both distirbutions should overlap considerably
print('mean train set accuracy: ', np.mean(clf['train_score']), ' +- ', np.std(clf['train_score']))
print('mean test set accuracy: ', np.mean(clf['test_score']), ' +- ', np.std(clf['test_score']))If we use Scikit-Learn, we need to use RandomSearchCV or GridSearchCV for hyperparameter tuning wth cross-validation. The code below is very similar to the previous one, but we pass our cross-validation scheme to GridSearchCV instead of cross_val_score().
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import (
KFold,
RepeatedKFold,
LeaveOneOut,
LeavePOut,
StratifiedKFold,
GridSearchCV, # RandomSearchCV
train_test_split,
)
# hyperparameter space
param_grid = dict(
penalty=['l1', 'l2'],
C=[0.1, 1, 10],
)
# K-Fold Cross-Validation
cv = KFold(n_splits=5, shuffle=True, random_state=4)
# Repeated K-Fold Cross-Validation
cv = RepeatedKFold(
n_splits=5,
n_repeats=10,
random_state=4,
)
# Leave One Out Cross-Validation
cv = LeaveOneOut()
# Leave P Out Cross-Validation
cv = LeavePOut(p=2)
# Leave P Out Cross-Validation
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=4)
# search
grid_search = GridSearchCV(
logit,
param_grid,
scoring='accuracy',
cv=cv, # any CV scheme, as defined in the previous section
refit=True, # refits best model to entire dataset
)
search = grid_search.fit(X_train, y_train)
# best hyperparameters
search.best_params_
# get all results
results = pd.DataFrame(search.cv_results_)[['params', 'mean_test_score', 'std_test_score']]The introduced cross-validation schemes were for independent and identically distirbuted (iid) data; however, some datasets don't fulfill the necessary requirements for that:
- Grouped data: multiple observations from the same subject; repeated measurements.
- Time series
For grouped data, the goal here would be to measure whether the model generalizes well for other subjects! To achieve that, the data from each subject is treated as a group and groups are moved around together; thus, we can use grouped K-Fold, or leave-one/p-out. In other words, groups are treated as data points before.
For time series, we want to predict future values. Thus, we create groups of time periods (e.g., months) and move them together. The additional constraint is that they are not shuffled and that if we predict month M, we use only the prior months as the train set.
See:
Example from 04-03-Group-Cross-Validation.ipynb, where we simulate repeated measurements of different patients:
# Logistic Regression
logit = LogisticRegression(
penalty ='l2', C=10, solver='liblinear', random_state=4, max_iter=10000)
# Group K-Fold Cross-Validation
# We are going to define groups as patient numbers
cv = GroupKFold(n_splits=5)
# Similarly, we have:
cv = LeaveOneGroupOut()
# estimate generalization error
clf = cross_validate(
logit,
X_train.drop('patient', axis=1), # drop the patient column, this is not a predictor
y_train,
scoring='accuracy',
return_train_score=True,
cv=cv.split(X_train.drop('patient', axis=1), y_train, groups=X_train['patient']),
)This topic is relevant for competitions.
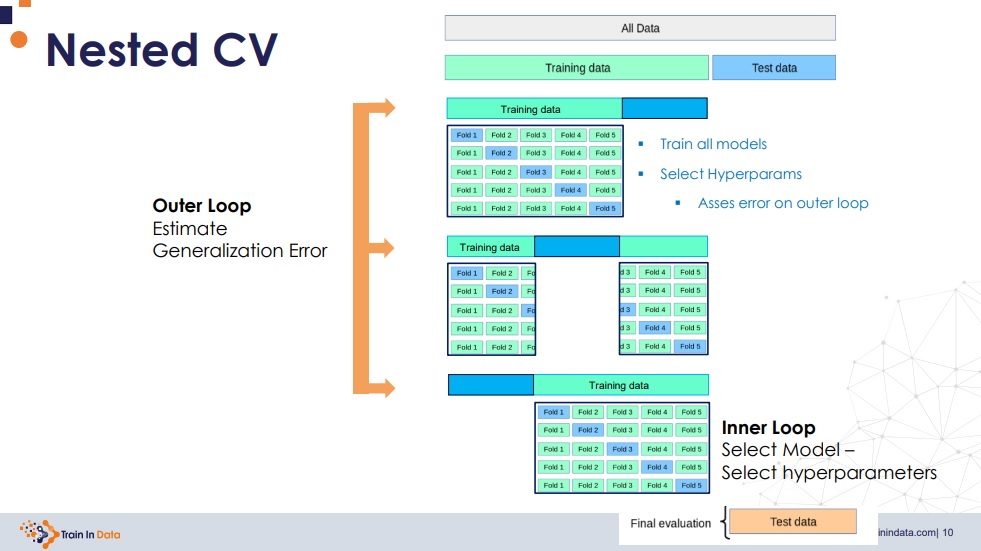
When we train different models with a hyperparameter search scheme (e.g., grid search) and a cross-validation scheme (e.g., K-fold), the generalization error is still positively biased. The reason is because in every k training, the validation subset is part of the training subest for another training; thus, we are leaking training information to the validation.
A solution to that consists in performing nested cross-validation: we perform a cross-validation within a cross-validation. This is an expensive approach, but used when we need a good generalization error estimation.
See notebook: 04-04-Nested-Cross-Validation.ipynb.
Things to consider:
- Number of hyperparameters of the machine learning model
- The low effective dimension: select hyperperameters that have an effect, and regions or spaces which are associated with changes in the response surface.
- The nature of the parameters: discrete, continuous, categorical; each type leads to different space definition startegies.
- The computing resources available to us: in general, the more combinations we try, the better, but that's not always true nor feasible.
We manually try with cross_val_score() different hyperparameter values and obtain the test errors (mean and standard deviation).
The idea is to obtain ranges of values that lead to models that generalize well, i.e., the error/score distirbutions of the train and test/val splits in the cross validation overlap and the unseen test split is contained in them.
When we have the approximate values, we define the ranges/values in a search scheme, e.g., GridSearchCV or RandomSearchCV.
Gris search is an exhaustive method: it tests all possible combinations of hyperparameter values we specify.
Grid search is very expensive, because the trials grow exponentially; however, it can be parallelized.
In practice, grid search is not enough, because it rarely finds the best sets of hyperparameters in the complete space. Instead, we use grid search for an initial search and then we fine tune with the results we obtain from it.
Important notebook, where all these concepts are implemented: 02-Grid-Search.ipynb. Nothing really new is introduced, but these ideas are correctly implemented:
- A first broad grid search is done and results are colledted in a data frame.
- The effect of each parameter is analyzed.
- The search space is narrowed down and grid search is applied again.
# set up the model
gbm = GradientBoostingClassifier(random_state=0)
# determine the hyperparameter space: 60 possible combinations
param_grid = dict(
n_estimators=[10, 20, 50, 100],
min_samples_split=[0.1, 0.3, 0.5],
max_depth=[1,2,3,4,None],
)
# set up the search
search = GridSearchCV(gbm, param_grid, scoring='roc_auc', cv=5, refit=True)
# find best hyperparameters
search.fit(X_train, y_train)
# get results
results = pd.DataFrame(search.cv_results_) # 60 x 16
# we can order the different models based on their performance
results.sort_values(by='mean_test_score', ascending=False, inplace=True)
results.reset_index(drop=True, inplace=True)
results[[
'param_max_depth', 'param_min_samples_split', 'param_n_estimators',
'mean_test_score', 'std_test_score',
]].head()
# plot model performance and error
results['mean_test_score'].plot(yerr=[results['std_test_score'], results['std_test_score']], subplots=True)
plt.ylabel('Mean test score')
plt.xlabel('Hyperparameter combinations')
# get overall generalization error
X_train_preds = search.predict_proba(X_train)[:,1]
X_test_preds = search.predict_proba(X_test)[:,1]
print('Train roc_auc: ', roc_auc_score(y_train, X_train_preds))
print('Test roc_auc: ', roc_auc_score(y_test, X_test_preds))
# Train roc_auc: 1.0
# Test roc_auc: 0.996766607877719
# let's make a function to evaluate the model performance based on
# single hyperparameters
def summarize_by_param(hparam):
tmp = pd.concat([
results.groupby(hparam)['mean_test_score'].mean(),
results.groupby(hparam)['mean_test_score'].std(),
], axis=1)
tmp.columns = ['mean_test_score', 'std_test_score']
return tmp
# check the effect of the parameter n_estimators
tmp = summarize_by_param('param_n_estimators')
tmp['mean_test_score'].plot(yerr=[tmp['std_test_score'], tmp['std_test_score']], subplots=True)
plt.ylabel('roc-auc')
# now repeat for all other parameters
# select the regions to refine in a new grid search
# determine the hyperparameter space
param_grid = dict(
n_estimators=[60, 80, 100, 120],
max_depth=[2,3],
loss = ['deviance', 'exponential'],
)
# set up the search
search = GridSearchCV(gbm, param_grid, scoring='roc_auc', cv=5, refit=True)
# find best hyperparameters
search.fit(X_train, y_train)
# the best hyperparameters are stored in an attribute
search.best_params_
# get all results
results = pd.DataFrame(search.cv_results_)
# perform comparisons/analysis if desired
# compute new generalization error, which should be better
X_train_preds = search.predict_proba(X_train)[:,1]
X_test_preds = search.predict_proba(X_test)[:,1]
print('Train roc_auc: ', roc_auc_score(y_train, X_train_preds))
print('Test roc_auc: ', roc_auc_score(y_test, X_test_preds))
# Train roc_auc: 0.9999999999999999
# Test roc_auc: 0.9973544973544973Note that in some cases a parameter value in a model enables other parameters. To deal with that, we can simply define 2+ dictionaries in the parameter grid; for instance, SVC:
# set up the model
svm = SVC(random_state=0)
# determine the hyperparameter space
param_grid = [
{'C': [1, 10, 100, 1000], 'kernel': ['linear']},
{'C': [1, 10, 100, 1000], 'gamma': [0.001, 0.0001], 'kernel': ['rbf']},
]
# set up the search
search = GridSearchCV(svm, param_grid, scoring='accuracy', cv=3, refit=True)
# find best hyperparameters
search.fit(X_train, y_train)While GridSearchCV explores all combinations, RandomSearchCV explores a randomly chosen subset from all possible combinations. It is very effective, because many parameters have regions with low effect. Therefore, if we have many hyperparameters, it is the preferred choice; if we have few hyperparameters, grid search is fine.
Random search is also well suited for continuous hyperparameters, because the search can draw values from a distribution; in contrast, grid search requires a manual definition of values to be tested. Therefore: instead of entering single values to RandomSearchCV, we should pass distirbutions to maximize its power!.
Important note: There is a probabilistic explanation that states that, independently of the search space, with 60 combinations we have a 95% probability of finding a set in neighborhood of the optimal set (top 5%). More on that: The "Amazing Hidden Power" of Random Search?.
For the rest, the usage of GridSearchCV and RandomSearchCV is quite similar.
from scipy import stats
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import (
RandomizedSearchCV,
train_test_split,
)
# set up the model
gbm = GradientBoostingClassifier(random_state=0)
# determine the hyperparameter space
# NOTE: we use distributions!
# To get random values:
# - integers: stats.randint.rvs(1, 5)
# - continuous/float: stats.uniform.rvs(0, 1)
# Here we don't call rvs() because we're ot drawing the numbers
# but passing the distribution
param_grid = dict(
n_estimators=stats.randint(10, 120),
min_samples_split=stats.uniform(0, 1),
max_depth=stats.randint(1, 5),
loss=('deviance', 'exponential'),
)
# set up the search
search = RandomizedSearchCV(gbm,
param_grid,
scoring='roc_auc',
cv=5,
n_iter = 60, # this is the number of combinations we want
random_state=10,
n_jobs=4,
refit=True)
# find best hyperparameters
search.fit(X_train, y_train)
# the best hyperparameters are stored in an attribute
search.best_params_
# we also find the data for all models evaluated
results = pd.DataFrame(search.cv_results_)
# we can order the different models based on their performance
results.sort_values(by='mean_test_score', ascending=False, inplace=True)
results.reset_index(drop=True, inplace=True)
results[[
'param_max_depth', 'param_min_samples_split', 'param_n_estimators',
'mean_test_score', 'std_test_score',
]].head()
# plot model performance and error
results['mean_test_score'].plot(yerr=[results['std_test_score'], results['std_test_score']], subplots=True)
plt.ylabel('Mean test score')
plt.xlabel('Hyperparameter combinations')
# generalization error
X_train_preds = search.predict_proba(X_train)[:,1]
X_test_preds = search.predict_proba(X_test)[:,1]
print('Train roc_auc: ', roc_auc_score(y_train, X_train_preds))
print('Test roc_auc: ', roc_auc_score(y_test, X_test_preds))
# let's make a function to evaluate the model performance based on
# single hyperparameters
def summarize_by_param(hparam):
tmp = pd.concat([
results.groupby(hparam)['mean_test_score'].mean(),
results.groupby(hparam)['mean_test_score'].std(),
], axis=1)
tmp.columns = ['mean_test_score', 'std_test_score']
return tmp
# performance change for n_estimators
tmp = summarize_by_param('param_n_estimators')
tmp['mean_test_score'].plot(yerr=[tmp['std_test_score'], tmp['std_test_score']], subplots=True)
plt.ylabel('roc-auc')Usually, we'll perform random searches with Scikit-Learn, but it's possible to do it with other packages, too, such as:
These packages were introduces to run Bayesian optimization, but they support also random search.
Notebooks where it is shown how:
Notebook: 05-Randomized-Search-with-Scikit-Optimize.ipynb
Equivalence to scikit-learn:
RandomSearchCVisdummy_minimize().- The hyperparameter space is defined in
param_gridusing specific type objects. - We define an
objective()function which gets theparam_gridvia a decorator; this function needs to manually computenp.mean(cross_val_score()).
import numpy as np
import pandas as pd
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import cross_val_score, train_test_split
from skopt import dummy_minimize # for the randomized search
from skopt.plots import plot_convergence
from skopt.space import Real, Integer, Categorical
from skopt.utils import use_named_args
# determine the hyperparameter space
# min, max, prior: distribution "uniform" (default) or "log-uniform", name
param_grid = [
Integer(10, 120, name="n_estimators"),
Real(0, 0.999, name="min_samples_split"),
Integer(1, 5, name="max_depth"),
Categorical(['deviance', 'exponential'], name="loss"),
]
# set up the gradient boosting classifier
gbm = GradientBoostingClassifier(random_state=0)
# We design a function to maximize the accuracy, of a GBM,
# with cross-validation
# the decorator allows our objective function to receive the parameters as
# keyword arguments. This is a requirement for scikit-optimize.
@use_named_args(param_grid)
def objective(**params):
# model with new parameters
gbm.set_params(**params)
# optimization function (hyperparam response function)
value = np.mean(
cross_val_score(
gbm,
X_train,
y_train,
cv=3,
n_jobs=-4,
scoring='accuracy')
)
# negate because we need to minimize
return -value
# Now, we could call objective() with a list of params
# to test it
# dummy_minimize performs the randomized search
search = dummy_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
n_calls=50, # the number of subsequent evaluations of f(x)
random_state=0,
)
# function value at the minimum.
# note that it is the negative of the accuracy
"Best score=%.4f" % search.fun # -0.9673
print("""Best parameters:
=========================
- n_estimators=%d
- min_samples_split=%.6f
- max_depth=%d
- loss=%s""" % (search.x[0],
search.x[1],
search.x[2],
search.x[3]))
plot_convergence(search)Note:
# To avoid an error I get with scikit-optimize
# I need to run these lines...
# https://stackoverflow.com/questions/63479109/error-when-running-any-bayessearchcv-function-for-randomforest-classifier
from numpy.ma import MaskedArray
import sklearn.utils.fixes
sklearn.utils.fixes.MaskedArray = MaskedArray
import skoptNotebook: 06-Randomized-Search-with-Hyperopt.ipynb
Equivalence to scikit-learn:
RandomSearchCVisfmin(); we specify the search algorithmrand- The hyperparameter space
param_gridis defined withhp. - A manually defined
objective()function needs to be passed tofmin();cross_val_score()needs to be used in it.
The hyperparameters are optimized for XGBoost; interesting links:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer
from sklearn.metrics import accuracy_score
from sklearn.model_selection import cross_val_score, train_test_split
import xgboost as xgb
from hyperopt import hp, rand, fmin, Trials
# hp: define the hyperparameter space
# rand: random search
# fmin: optimization function
# Trials: to evaluate the different searched hyperparameters
# load dataset
breast_cancer_X, breast_cancer_y = load_breast_cancer(return_X_y=True)
X = pd.DataFrame(breast_cancer_X)
y = pd.Series(breast_cancer_y).map({0:1, 1:0})
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0)
# determine the hyperparameter space
# http://hyperopt.github.io/hyperopt/getting-started/search_spaces/
# NOTE: q-distributions return a float, thus, sometimes recasting needs to be done!
# hp.choice: returns one of several options (suitable for categorical hyperparams)
# hp.randint: returns a random integer between 0 and an upper limit
# hp.uniform: returns a value uniformly between specified limits
# hp.quniform: Returns a value like round(uniform(low, high) / q) * q, i.e. sample between min and max in steps of size q
# hp.loguniform: draws values from exp(uniform(low, high)) so that the logarithm of the returned value is uniformly distributed
# hp.qloguniform: Returns a value like round(exp(uniform(low, high)) / q) * q (similar use and cautions to hp.quniform but for log-uniform distributions)
# hp.normal: draws from a normal distribution with specified mu and sigma
# hp.qnormal: Returns a value like round(normal(mu, sigma) / q) * q
# hp.lognormal: Returns a value drawn according to exp(normal(mu, sigma)) so that the logarithm of the return value is normally distributed
# hp.qlognormal: Returns a value like round(exp(normal(mu, sigma)) / q) * q
param_grid = {
'n_estimators': hp.quniform('n_estimators', 200, 2500, 100),
'max_depth': hp.uniform('max_depth', 1, 10), # min, max
'learning_rate': hp.uniform('learning_rate', 0.01, 0.99),
'booster': hp.choice('booster', ['gbtree', 'dart']),
'gamma': hp.quniform('gamma', 0.01, 10, 0.1),
'subsample': hp.uniform('subsample', 0.50, 0.90),
'colsample_bytree': hp.uniform('colsample_bytree', 0.50, 0.99),
'colsample_bylevel': hp.uniform('colsample_bylevel', 0.50, 0.99),
'colsample_bynode': hp.uniform('colsample_bynode', 0.50, 0.99),
'reg_lambda': hp.uniform('reg_lambda', 1, 20)
}
# the objective function takes the hyperparameter space
# as input
def objective(params):
# we need a dictionary to indicate which value from the space
# to attribute to each value of the hyperparameter in the xgb
# here, we capture one parameter from the distributions
params_dict = {
'n_estimators': int(params['n_estimators']), # important int, as it takes integers only
'max_depth': int(params['max_depth']), # important int, as it takes integers only
'learning_rate': params['learning_rate'],
'booster': params['booster'],
'gamma': params['gamma'],
'subsample': params['subsample'],
'colsample_bytree': params['colsample_bytree'],
'colsample_bylevel': params['colsample_bylevel'],
'colsample_bynode': params['colsample_bynode'],
'random_state': 1000,
}
# with ** we pass the items in the dictionary as parameters
# to the xgb
gbm = xgb.XGBClassifier(**params_dict)
# train with cv
score = cross_val_score(gbm, X_train, y_train,
scoring='accuracy', cv=5, n_jobs=4).mean()
# to minimize, we negate the score
return -score
# Now, we could call objective() with a list of params
# to test it
# OPTIONAL: We can use the Trials() object to store more information from the search
# for later inspection
trials = Trials()
# fmin performs the minimization
# rand.suggest samples the parameters at random
# i.e., performs the random search
# NOTE: I got the error "AttributeError: 'numpy.random._generator.Generator' object has no attribute 'randint'"
# So I needed to replace randint() with integers() in fmin.py
search = fmin(
fn=objective,
space=param_grid,
max_evals=50, # number of combinations we'd like to randomly test
rstate=np.random.default_rng(42),
algo=rand.suggest, # randomized search
trials=trials
)
# best hyperparameters
search
# the best hyperparameters can also be found in
# trials
trials.argmin
# All the results, sorted by loss = minimized score
results = pd.concat([
pd.DataFrame(trials.vals),
pd.DataFrame(trials.results)],
axis=1,
).sort_values(by='loss', ascending=False).reset_index(drop=True)
# Plot the score as function of the hyperparameter combination
results['loss'].plot()
plt.ylabel('Accuracy')
plt.xlabel('Hyperparam combination')
# Minimum/best score (recall we minimize)
pd.DataFrame(trials.results)['loss'].min()
# create another dictionary to pass the search items as parameters
# to a new xgb
# we basically grab the best hyperparameters here and assure that the types
# are correct
best_hp_dict = {
'n_estimators': int(search['n_estimators']), # important int, as it takes integers only
'max_depth': int(search['max_depth']), # important int, as it takes integers only
'learning_rate': search['learning_rate'],
'booster': 'gbtree',
'gamma': search['gamma'],
'subsample': search['subsample'],
'colsample_bytree': search['colsample_bytree'],
'colsample_bylevel': search['colsample_bylevel'],
'colsample_bynode': search['colsample_bynode'],
'random_state': 1000,
}
# after the search we can train the model with the
# best parameters manually
gbm_final = xgb.XGBClassifier(**best_hp_dict)
gbm_final.fit(X_train, y_train)
# Final score on train and test splits
X_train_preds = gbm_final.predict(X_train)
X_test_preds = gbm_final.predict(X_test)
print('Train accuracy: ', accuracy_score(y_train, X_train_preds))
print('Test accuracy: ', accuracy_score(y_test, X_test_preds))Recall that we have a response surface Phi() which depends on the hyperparameters lambda; that response surface is the metric we want to optimize. We don't have a closed form of the response surface function, instead it's a black box.
Black box optimization can be performed with GridSearchCV and RandomSearchCV when we have simple models, i.e., when the cost of evaluating Phi is low; we can do it even in parallel. If we are training neural networks, that's not the case: it's not possible to train many combinations of hyperparameters.
For neural networks, sequential searches are better suited: we try a set and then we compute which values we'd need to change to try again. That needs to be carried out in sequence, and the cost of computing the new values needs to be lower than evaluating the model. Bayesian optimization is one of the sequential search methods used for black box optimization; it makes sense using it when the black box objective function (the model) is costly to evaluate.
To use Bayesian optimization, the optimized objective function f must be:
- f is continuous
- f is difficult to evaluate; too much time or money
- f lacks known structure, like concavity or linearity; f is black-box
- f has no derivative; we can’t evaluate a gradient
- f can be evaluated at arbitrary points of x (the hyperparameters)
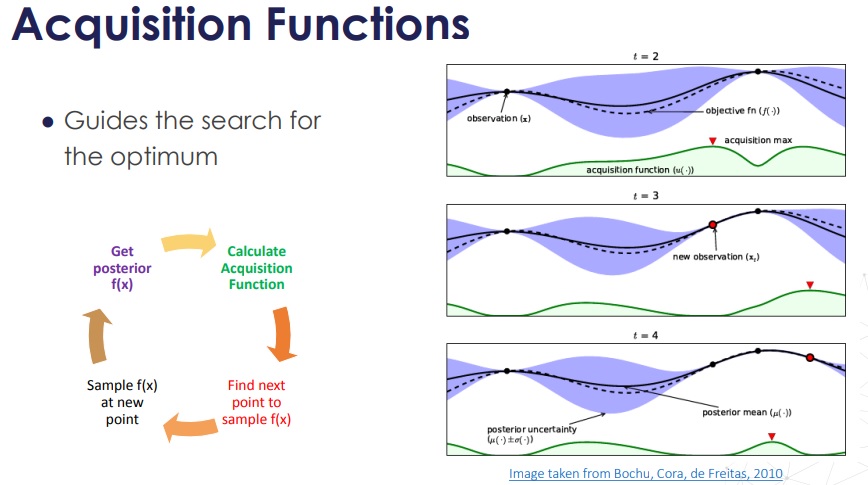
In Bayesian optimization, we follow these steps:
- In Bayesian optimization we treat f as a random function and place a prior over it. The prior is a function that captures the belief (distribution, behaviour) of f. To estimate the prior, we can use
- Gaussian processes
- Tree-parzen estimator
- Random Forests
- Then, we evaluate f at certain points: we evaluate the model with some hyperparameter combinations.
- With the new data, the prior (f original belief) is updated to a new the posterior distribution.
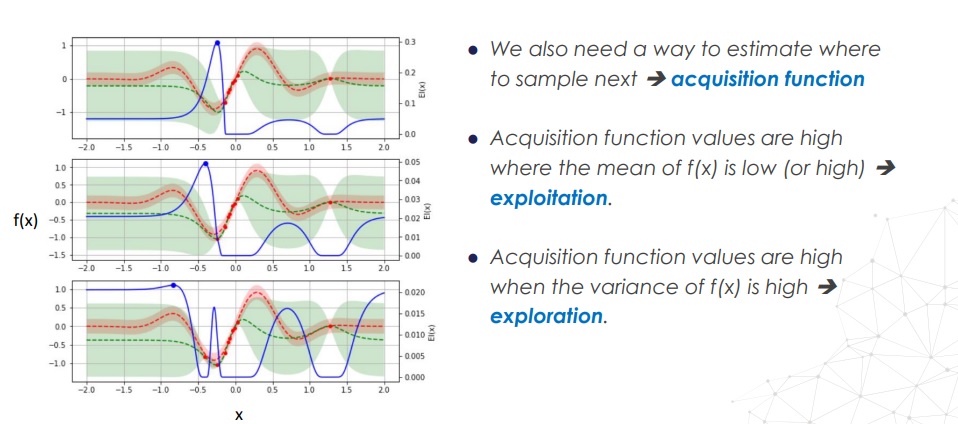
- The posterior distribution is used to construct an acquisition function to determine the next query point (i.e., which new hyperparameters to use next in step 2). The acquisition function can be
- Expected Improvement (EI)
- Gaussian process upper confidence bound (UCB)
In Bayesian inference, we have:
- A prior unconditonal belief which is a distribution pointing the probability of some variables.
- A posterior conditional belief, which is the updated prior belief after we have gathered more information. The posterior belief is also a distirbution of the probabilities of some variables.
The step to go from the prior to the posterior is done with the Bayes rule; the posterior becomes the new prior in the next iteration.
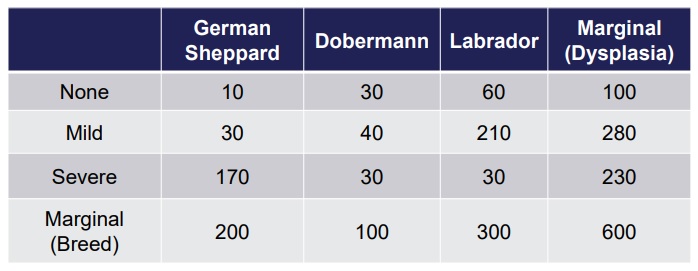
Example: we have a table with dog breeds and number of dogs which have dysplasia of a given degree.
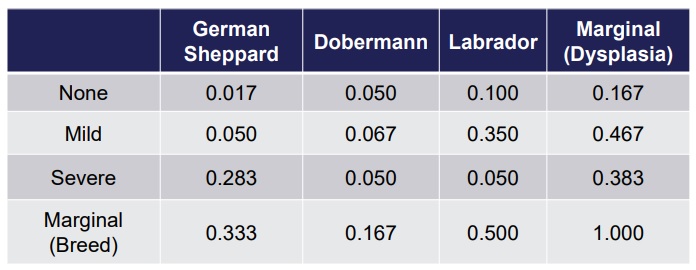
We can compute the frequencies dividing by the total number of dogs:
Definitions of probabilities:
- Marginal probability:
P(A), P(B): probability of a variable:P(Breed = Labrador) = 0.5,P(Dysplasia = Mild) = 0.467. - Joint probability:
P(A,B): any cell in the table, i.e., the probability that the variables take a given value:P(Breed = Labrador, Dysplasia = Mild) = 0.35.- It is symmetric:
P(A,B) = P(B,A). - The marginal probability is the sum of joint probabilities:
P(A) = sum(P(A,B), for all B categories).
- It is symmetric:
- Conditional probability:
P(A|B): we fix the value of a variable and compute the probabilities of the categories of the other variable, e.g., we know a dog is a Labrador, which is the probability of it having a mild dysplasia?P(Dysplasia = Mild | Breed = Labrador).- Computation:
P(A|B) = P(A,B) / P(B),P(Dysplasia = Mild | Breed = Labrador) = P(Dysplasia = Mild, Breed = Labrador) / P(Breed = Labrador) = 0.35 / 0.5 = 0.7 - It is not symmetric!
P(A|B) != P(B|A)
- Computation:
From the above, the Bayes rule is derived:
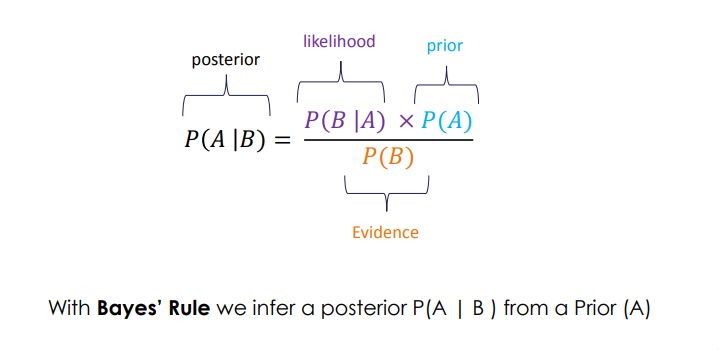
P(A|B) = P(B|A) * P(A) / P(B)
The nice thing of the Bayes rule is that we can use it to obtain a better estimate (P(A|B)) of a prior belief (P(A)) given some evidence (P(B)).
The basic idea of Bayesian optimization for hyperparameter tuning is the following:
P(A) -> A: f(x), i.e., the objective function, the model performance.x: the vector with the hyperparameters.
P(B) -> B: Data, the available dataP(Data): the denominator can be dropped, since it only scales.- We don't take the real
f(x), but take a surrogate functionf(x)which is a multivariate Gaussian distribution, called Gaussian Process; this surrogate function is like an approximation or best estimate of our objective function. For each pointx(the vector of hyperparamters), we have an estimated mean offand a covariance matrix ofxwhich captures the uncertainty as a spread. In the beginning, the surrogate Gaussian Process provides a constant mean value with a wide spread, no matter the vector value forx. The idea is to refine that surrogatefby evaluating the real model and to find the optimum of the surrogate, which will capture the optimum of the realf(x). - We evaluate our model with some values of
x = x_0; at these point, the value off(x = x_0)becomes known, the spread decreases around it. - The goal is to find the minimum of that surrogate
f. In essence the problem is a regression problem (hence, we need a Gaussian Process regressor), but instead of discovering the shape off, we are interested in discovering only its optimum. - We define an acquisition function, which has high values in areeas which
fmight have a minimum (more on this later). - Following the acquisition function, we pick the next
x_1and evaluate the model there, obtaining a better estimate of the surrogate function. - The new surrogate function updates the acquisition function, which leads to a new point.
- At the end, we converge to a minimum of the surrogate
f, which approximates our model!
In order to model the spread or uncertainty of the Gaussian Process or surrogate f(x), we use the covariance matrix of the hyperparameters x. That covariance matrix is composed by kernels: functions that depend on a distance norm between two x variables. The most common kernels are:
- Exponential or Radial Basis Function (RBF):
k(x_i, x_j) = alpha * exp(-((x_i - x_j)^2)/2*s^2) - Martérn: a function of Gamma and Bessel functions; used by default by
gp_minimizefrom Scikit-Optimize.
There are several choices for acquisition functions:
- Probability of Improvement (PI): Probability that the new sampled value is bigger than the highest observed value. It is computed by the cummulative probability of an
f(x)distribution above the maximum seen value. - Expected Improvement (EI): better that the previous.
- Upper (or Lower) confidence bound (UCB or LCB): we add of substract the spread to the mean of
f(x)to compute the lower/upper estimates off(x).
Also, note that there is a trade-off decision inherent to the acquisition function, which depends on the exploration vs. exploitation strategy which is taken; we might want to refine a probably minimum of the function, but with that we don't explore unkown regions with high uncertainty.
- Lecture by Nando de Freitas: Machine learning - Introduction to Gaussian processes
- A Tutorial on Bayesian Optimization of Expensive Cost Functions
- Taking the Human Out of the Loop: A Review of Bayesian Optimization
- A Tutorial on Bayesian Optimization
- Practical Bayesian Optimization of Machine Learning Algorithms
- Bayesian Optimization Primer
The rest of this section focuses on Scikit-Optimize, aka. skopt, for Bayesian optimization of tree-based models and neural networks.
There are two main APIs for Bayesian optimization in skopt:
- The manual API, in which we define the
objective()function which performs the model training given a set of hyperparameters. Here, we can use any model and any cross-validation scheme. For instance, we can use scikit-learn models withcross_val_score(). Theobjective()function is passed togp_minimize(), which performs the Bayesian optimization. We can also change the kernels and tune theGaussianProcessRegressor. The manual API is more complex, but provides a lot of flexibility, so that we can optimize any model of any ML framework with it! - The automatic API, which mimicks the Scikit-Learn objects: we instantiate
BayesSearchCVand pass to it the hyperparameter space and asklearn-conform estimator; then, everything is done automatically, as it were aGridSearchCVobject.
Notes:
- Bayesian optimization is not only for hyperparameter tuning; we can use it with any function which is hard to evaluate and of which we don't have a closed-form formula.
- Bayesian optimization is costly; it makes sense when the model we're training is costly to train and evaluate, e.g., a complex XGBoost model or neural networks. Otherwise,
GridSearchCV, or betterRandomSearchCVwith distirbutions and 60 calls should be used. - Scikit-optimize is very sensitive to the Scikit-Learn and Scipy versions used; additionally, it seems there are no new versions of Scikit-Optimize in the past 2 years. Sometimes some examples from the course yield errors, probably due to version conflicts. Used versions:
numpy==1.22.0 scipy==1.5.3 scikit-learn==0.23.2 scikit-optimize==0.8.1 - Maybe an important remark is that Scikit-Optimize is currently not updated/maintained regularly; the last release to date (2023-05) is v0.9 in 2021-10.
- Always plot the convergence with
plot_convergence(). We can see how many iterations are really necessary, and whether we reached the optimum; even though we might usen_calls=50often times, sometimes30calls is more than enough. - To analyze the optimization process (i.e., response as function of hyperparameters and hyperparameter values in time), use, respectively:
plot_objective()andplot_evaluations().
The most important notebooks/sub-sections:
04-Bayesian-Optimization-GBM-Grid.ipynb: Automatic Bayesian optimization of asklearnGBT usingBayesSearchCV.06-Bayesian-Optimization-xgb-Optional.ipynb: Manual Bayesian optimization of an XGBoost classifier.07-Bayesian-Optimization-CNN.ipynb: Manual Bayesian optimization of a TF/Keras-CNN.
In the notebook 01-Bayesian-Optimization-Demo.ipynb, the minimum of an unkown 1D function is found using Gaussian processes with scikit-optimize; the function is treated as unknown (i.e., black box, no close form), but we can evaluate it and any point.
The example shows that Bayesian optimization is not only for hyperparameter tuning, but for any optimization of black box functions!
Notebook: 02-Bayesian-Optimization-1-Dimension.ipynb.
A Gradient Boosted Tree is tuned using Bayes optimization for one hyperparameter (the number of nodes). It doesn't make sense to use Gaussian process optimization (Bayes optimization) for a tree with one hyperparameter, but this notebook is for demo purposes.
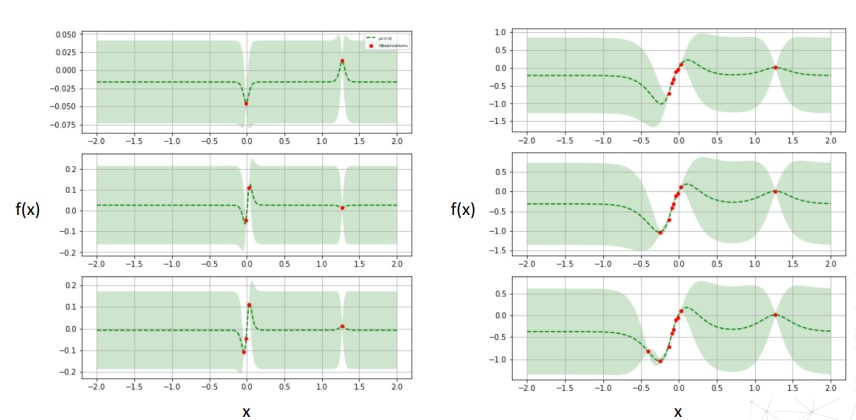
Apart from performing the optimization, the Gaussian process modification is plotted in different stages (including the acquisition function values).
Note: I needed to fix a bug in the skopt package in the file gpr.py; the code was bifurcating depending on the sklearn minor version.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import (
cross_val_score,
train_test_split,
GridSearchCV,
)
from skopt import gp_minimize
from skopt.plots import plot_convergence, plot_gaussian_process
from skopt.space import Integer
from skopt.utils import use_named_args
# set up the model
gbm = GradientBoostingClassifier(random_state=0)
# determine the hyperparameter space
param_grid = {
'n_estimators': [10, 20, 50, 100, 120, 150, 200, 250, 300],
}
# set up the search
# we perform a grid search to know the shape of the response surface
# which will be estimated later in the Gaussian process optimization
search = GridSearchCV(gbm, param_grid, scoring='accuracy', cv=3, refit=False)
# find best hyperparameter
search.fit(X_train, y_train)
results = pd.DataFrame(search.cv_results_)
results.sort_values(by='param_n_estimators', ascending=False, inplace=True)
# plot f(x) - 1-D hyperparameter response function
plt.plot(results['param_n_estimators'], results['mean_test_score'], "ro--")
plt.ylabel('Accuracy')
plt.xlabel('Number of trees')
# Now, we perform Bayesian optimization
# The first step consists in defining the hyperparameter space
# More info: https://scikit-optimize.github.io/stable/modules/generated/skopt.Space.html
# min, max, prior: distribution "uniform" (default) or "log-uniform", name
param_grid = [Integer(10, 300, name="n_estimators")]
# We design a function to maximize the accuracy, of a GBM,
# with cross-validation
# the decorator allows our objective function to receive the parameters as
# keyword arguments. This is a requirement of Scikit-Optimize.
@use_named_args(param_grid)
def objective(**params):
# model with new parameters
gbm.set_params(**params)
# optimization function (hyperparam response function)
value = np.mean(
cross_val_score(
gbm,
X_train,
y_train,
cv=3,
n_jobs=-4,
scoring='accuracy')
)
# negate metric because we need to minimize
return -value
# Now, we could call objective() with a list of params
# to test it
# gp_minimize performs by default GP Optimization
# https://scikit-optimize.github.io/stable/modules/generated/skopt.gp_minimize.html
gp_ = gp_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
n_initial_points=2, # the number of points to evaluate f(x) to start of
acq_func='EI', # the acquisition function
n_calls=20, # the number of subsequent evaluations of f(x)
random_state=0,
)
# we can access the hyperparameter space
gp_.space
# function value at the minimum.
# note that it is the negative of the accuracy
"Best score=%.4f" % gp_.fun
print("""Best parameters:
=========================
- n_estimators=%d""" % (gp_.x[0]))
# Plot the score during the steps.
# This shows whether we have converged or not, i.e., whether we finished or not.
# We can see that the minimum is found very fast
# almost in the 2nd call
# https://scikit-optimize.github.io/stable/modules/generated/skopt.plots.plot_convergence.html#skopt.plots.plot_convergence
plot_convergence(gp_)
# The folloing code snippet shows the different steps
# of the Gaussian optimization with their values:
# - in green the surrogate f(x) which is being discovered and minimized
# - in orange the precomputed response surface f(x) (with grid search)
# - in red the trials/evaluations, i.e., the observations
# - in blue the acquisition function EI
# https://scikit-optimize.github.io/stable/modules/generated/skopt.plots.plot_gaussian_process.html#skopt.plots.plot_gaussian_process
end = 10
for n_iter in range(1, end):
# figure size
plt.figure(figsize=(10,20))
# ===================
# 2 plots next to each other, left plot
plt.subplot(end, 2, 2*n_iter+1)
# plot gaussian process search
ax = plot_gaussian_process(
gp_,
n_calls=n_iter,
show_legend=True,
show_title=False,
show_next_point=False,
show_acq_func=False)
# plot true hyperparameter response function
ax.scatter(results['param_n_estimators'], -results['mean_test_score'])
ax.set_ylabel("")
ax.set_xlabel("")
# ===================
# Plot EI(x) - the acquisition function
# 2 plots next to each other, right plot
plt.subplot(end, 2, 2*n_iter+2)
ax = plot_gaussian_process(
gp_,
n_calls=n_iter,
show_legend=True,
show_title=False,
show_mu=False,
show_acq_func=True,
show_observations=False,
show_next_point=True)
ax.set_ylabel("")
ax.set_xlabel("")
plt.show()Note:
# To avoid an error I get with scikit-optimize
# I need to run these lines...
# https://stackoverflow.com/questions/63479109/error-when-running-any-bayessearchcv-function-for-randomforest-classifier
from numpy.ma import MaskedArray
import sklearn.utils.fixes
sklearn.utils.fixes.MaskedArray = MaskedArray
import skoptNotebook: 04-Bayesian-Optimization-GBM-Manual.ipynb.
This notebook is very similar to the previous one, but we optimize 4 hyperparameters in a gradient boosted tree.
Unfortunately, I get an error when running the code. I have tried to install different versions of scikit-learn ans scikit-optimize, but the error persists.
However, the error is not that important, because
- in the next subsection/notebook,
BayesSearchCVis introduced, which works and resembles theGridSearchCVAPI from Scikit-Learn; - in the subsection after that, a change of kernel is performed and this manual approach works;
- in the subsection after that an XGBoost model (equivalent to the GBM) is optimized manually without problems.
from skopt.space import Real, Integer, Categorical
# The only difference in the code is the definition
# of the hyperparameter space
# min, max, prior: distribution "uniform" (default) or "log-uniform", name
param_grid = [
Integer(10, 120, name="n_estimators"),
Real(0.0001, 0.999, name="min_samples_split"),
Integer(1, 5, name="max_depth"),
Categorical(['log_loss', 'exponential'], name="loss"),
]Example: Automatic Gaussian Optimization of a Grandient Boosted Tree with 4 Hyperparameters (BayesSearchCV)
Notebook: 04-Bayesian-Optimization-GBM-Grid.ipynb.
This notebook is very similar to the previous one: we optimize 4 hyperparameters in a gradient boosted tree, but using BayesSearchCV, which resembles the GridSearchCV API from Scikit-Learn.
Important note: I think this is the easiest way of performing hyperparameter optimization using Bayes for models from Scikit-Learn.
Also, note that I didn't get an error when running this notebook.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy import stats
from sklearn.datasets import load_boston
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
# note that we only need to import the wrapper
from skopt import BayesSearchCV
# load dataset
boston_X, boston_y = load_boston(return_X_y=True)
X = pd.DataFrame(boston_X)
y = pd.Series(boston_y)
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0)
# set up the model
gbm = GradientBoostingRegressor(random_state=0)
# hyperparameter space
# now, it is a dictionary (not a list as in the manual case),
# beause BayesSearchCV tries to mimick scikit-learn
param_grid = {
'n_estimators': (10, 120), # min, max
'min_samples_split': (0.001, 0.99, 'log-uniform'), # min, max, sample uniformly in log scale
'max_depth': (1, 8), # min, max
'loss': ['ls', 'lad', 'huber'], # list of classes
}
# set up the search
search = BayesSearchCV(
estimator=gbm,
search_spaces=param_grid,
scoring='neg_mean_squared_error', # same as in scikit-learn
cv=3,
n_iter=50,
random_state=10,
n_jobs=4, # number of processors
refit=True)
# find best hyperparameters
search.fit(X_train, y_train)
# the best hyperparameters are stored in an attribute
search.best_params_
# the best hyperparameters are stored in an attribute
search.best_params_
# OrderedDict([('loss', 'huber'),
# ('max_depth', 3),
# ('min_samples_split', 0.001),
# ('n_estimators', 101)])
# the best score
search.best_score_ # -11.292655185099154
# we also find the data for all models evaluated
results = pd.DataFrame(search.cv_results_)
# we can order the different models based on their performance
results.sort_values(by='mean_test_score', ascending=False, inplace=True)
results.reset_index(drop=True, inplace=True)
# plot model performance and error
results['mean_test_score'].plot(yerr=[results['std_test_score'], results['std_test_score']], subplots=True)
plt.ylabel('Mean test score')
plt.xlabel('Hyperparameter combinations')
# Generalization error
X_train_preds = search.predict(X_train)
X_test_preds = search.predict(X_test)
print('Train MSE: ', mean_squared_error(y_train, X_train_preds))
print('Test MSE: ', mean_squared_error(y_test, X_test_preds))Notebook: 05-Bayesian-Optimization-Change-Kernel.ipynb
By default, the kernel used by the Gaussian Process regressor is Martérn. However, we can change it to be, e.g., the RBF (exponential) kernel.
In that case, we need to use the manual API, in which we define an objective function with a decorator; additionally, we need to define a new GaussianProcessRegressor which is passed to gp_minimize().
Apart from that, the code is very similar.
Links:
import numpy as np
import pandas as pd
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import cross_val_score, train_test_split
# squared exponential kernel
from sklearn.gaussian_process.kernels import RBF
from skopt import gp_minimize
from skopt.plots import plot_convergence
from skopt.space import Real, Integer, Categorical
from skopt.utils import use_named_args
# Gaussian Process Regressor, we will change the kernel here:
from skopt.learning import GaussianProcessRegressor
# load dataset
breast_cancer_X, breast_cancer_y = load_breast_cancer(return_X_y=True)
X = pd.DataFrame(breast_cancer_X)
y = pd.Series(breast_cancer_y).map({0:1, 1:0})
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0)
# determine the hyperparameter space
# min, max, prior: distribution "uniform" (default) or "log-uniform", name
param_grid = [
Integer(10, 120, name="n_estimators"),
Real(0, 0.999, name="min_samples_split"),
Integer(1, 5, name="max_depth"),
Categorical(['deviance', 'exponential'], name="loss"),
]
# set up the gradient boosting classifier
gbm = GradientBoostingClassifier(random_state=0)
# define the kernel: RBF (exponential)
# https://scikit-learn.org/stable/modules/generated/sklearn.gaussian_process.kernels.RBF.html#sklearn.gaussian_process.kernels.RBF
kernel = 1.0 * RBF(length_scale=1.0, length_scale_bounds=(1e-1, 10.0))
# Set up manually the GaussianProcessRegressor
# with the kernel
gpr = GaussianProcessRegressor(
kernel=kernel,
normalize_y=True, noise="gaussian",
n_restarts_optimizer=2
)
# We design a function to maximize the accuracy, of a GBM,
# with cross-validation
# the decorator allows our objective function to receive the parameters as
# keyword arguments. This is a requirement of Scikit-Optimize.
@use_named_args(param_grid)
def objective(**params):
# model with new parameters
gbm.set_params(**params)
# optimization function (hyperparam response function)
value = np.mean(
cross_val_score(
gbm,
X_train,
y_train,
cv=3,
n_jobs=-4,
scoring='accuracy')
)
# negate because we need to minimize
return -value
# Now, we could call objective() with a list of params
# to test it
# Bayesian optimization
gp_ = gp_minimize(
objective,
dimensions=param_grid,
base_estimator=gpr,
n_initial_points=5,
acq_optimizer="sampling",
random_state=42,
)
# we can access the hyperparameter space
gp_.space
# function value at the minimum.
# note that it is the negative of the accuracy
"Best score=%.4f" % gp_.fun
# 'Best score=-0.9724'
print("""Best parameters:
=========================
- n_estimators=%d
- min_samples_split=%.6f
- max_depth=%d
- loss=%s""" % (gp_.x[0],
gp_.x[1],
gp_.x[2],
gp_.x[3]))
# Best parameters:
# =========================
# - n_estimators=119
# - min_samples_split=0.764091
# - max_depth=4
# - loss=exponential
# Convergence plot
# https://scikit-optimize.github.io/stable/modules/generated/skopt.plots.plot_convergence.html#skopt.plots.plot_convergence
plot_convergence(gp_)Notebook: 06-Bayesian-Optimization-xgb-Optional.ipynb.
In this notebook, instead of optimizing the hyperparameters of a Gradient Boosted Tree from Scikit-Learn, an equivalent XGBoost model is tuned. In contrast to the GBT, this time the optimization takes place without any errors.
This notebook, together with the BayesSearchCV and the next Keras-CNN notebook is the summary of practical usage of Bayesian optimization with Scikit-Optimize.
import numpy as np
import pandas as pd
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import cross_val_score, train_test_split
import xgboost as xgb
from skopt import gp_minimize
from skopt.plots import plot_convergence
from skopt.space import Real, Integer, Categorical
from skopt.utils import use_named_args
# load dataset
breast_cancer_X, breast_cancer_y = load_breast_cancer(return_X_y=True)
X = pd.DataFrame(breast_cancer_X)
y = pd.Series(breast_cancer_y).map({0:1, 1:0})
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0)
# determine the hyperparameter space
# min, max, prior: distribution "uniform" (default) or "log-uniform", name
param_grid = [
Integer(200, 2500, name='n_estimators'),
Integer(1, 10, name='max_depth'),
Real(0.01, 0.99, name='learning_rate'),
Categorical(['gbtree', 'dart'], name='booster'),
Real(0.01, 10, name='gamma'),
Real(0.50, 0.90, name='subsample'),
Real(0.50, 0.90, name='colsample_bytree'),
Real(0.50, 0.90, name='colsample_bylevel'),
Real(0.50, 0.90, name='colsample_bynode'),
Integer(1, 50, name='reg_lambda'),
]
# set up the gradient boosting classifier
gbm = xgb.XGBClassifier(random_state=1000)
# We design a function to maximize the accuracy, of a GBM,
# with cross-validation
# the decorator allows our objective function to receive the parameters as
# keyword arguments. This is a requirement of Scikit-Optimize.
@use_named_args(param_grid)
def objective(**params):
# model with new parameters
gbm.set_params(**params)
# optimization function (hyperparam response function)
value = np.mean(
cross_val_score(
gbm,
X_train,
y_train,
cv=3,
n_jobs=-4,
scoring='accuracy')
)
# negate because we need to minimize
return -value
# Before we run the hyper-parameter optimization,
# let's first check that the everything is working
# by passing some default hyper-parameters.
default_parameters = [200,
3,
0.1,
'gbtree',
0.1,
0.6,
0.6,
0.6,
0.6,
10]
objective(x=default_parameters)
# -0.9598048150679729
# gp_minimize performs by default GP Optimization
# using a Marten Kernel
# NOTE: in the case of neural networks, instead of passing
# n_initial_points, it is common to pass the default/initial hyperparams
# in the argument x0
gp_ = gp_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
n_initial_points=10, # the number of points to evaluate f(x) to start of
acq_func='EI', # the acquisition function
n_calls=50, # the number of subsequent evaluations of f(x)
random_state=0,
)
# we can access the hyperparameter space
gp_.space
# function value at the minimum.
# note that it is the negative of the accuracy
"Best score=%.4f" % gp_.fun
print("""Best parameters:
=========================
- n_estimators = %d
- max_depth = %d
- learning_rate = %.6f
- booster = %s
- gamma = %.6f
= subsample = %.6f
- colsample_bytree = %.6f
- colsample_bylevel = %.6f
- colsample_bynode' = %.6f
""" % (gp_.x[0],
gp_.x[1],
gp_.x[2],
gp_.x[3],
gp_.x[4],
gp_.x[5],
gp_.x[6],
gp_.x[7],
gp_.x[8],
))
# Plot convergence
# https://scikit-optimize.github.io/stable/modules/generated/skopt.plots.plot_convergence.html#skopt.plots.plot_convergence
plot_convergence(gp_)Notebook: 07-Bayesian-Optimization-CNN.ipynb.
Very interesting notebook, in which Bayesian optimization is applied to a CNN model defined with Tensorflow/Keras. The dataset with which it is trained is MNIST Kaggle.
New analysis plots are introduced:
plot_objective(): it shows the response as function of different hyperparameter values.plot_evaluations(): it shows the selection of hyperparameter values along the different calls.
# For reproducible results.
# See:
# https://keras.io/getting_started/faq/#how-can-i-obtain-reproducible-results-using-keras-during-development
import os
os.environ['PYTHONHASHSEED'] = '0'
import numpy as np
import tensorflow as tf
import random as python_random
# The below is necessary for starting Numpy generated random numbers
# in a well-defined initial state.
np.random.seed(123)
# The below is necessary for starting core Python generated random numbers
# in a well-defined state.
python_random.seed(123)
# The below set_seed() will make random number generation
# in the TensorFlow backend have a well-defined initial state.
# For further details, see:
# https://www.tensorflow.org/api_docs/python/tf/random/set_seed
tf.random.set_seed(1234)
import itertools
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from keras.utils.np_utils import to_categorical
from keras.models import Sequential, load_model
from keras.layers import Dense, Flatten, Conv2D, MaxPool2D
from keras.optimizers import Adam
from keras.callbacks import ReduceLROnPlateau
from skopt import gp_minimize
from skopt.space import Real, Categorical, Integer
from skopt.plots import plot_convergence
from skopt.plots import plot_objective, plot_evaluations
from skopt.utils import use_named_args
# Load the data
data = pd.read_csv("../data/train.csv")
# first column is the target, the rest of the columns
# are the pixels of the image
# each row is 1 image
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
data.drop(['label'], axis=1), # the images
data['label'], # the target
test_size = 0.1,
random_state=0)
X_train.shape, X_test.shape
# ((37800, 784), (4200, 784))
# Re-scale the data
# 255 is the maximum value a pixel can take
X_train = X_train / 255
X_test = X_test / 255
# Reshape image in 3 dimensions:
# height: 28px X width: 28px X channel: 1
X_train = X_train.values.reshape(-1,28,28,1)
X_test = X_test.values.reshape(-1,28,28,1)
# the target is 1 variable with the 9 different digits
# as values
y_train.unique()
# 2, 0, 7, 4, 3, 5, 9, 6, 8, 1
# For Keras, we need to create 10 dummy variables,
# one for each digit
# Encode labels to one hot vectors (ex : digit 2 -> [0,0,1,0,0,0,0,0,0,0])
y_train = to_categorical(y_train, num_classes = 10)
y_test = to_categorical(y_test, num_classes = 10)
# Some image examples
g = plt.imshow(X_train[0][:,:,0])
# function to create the CNN
# we pass the hyperparameters we want to optimize
# and the function returns the model (compiled with the optimizer)
# inside the function the model is defined
def create_cnn(
learning_rate,
num_dense_layers,
num_dense_nodes,
activation,
):
"""
Hyper-parameters:
learning_rate: Learning-rate for the optimizer.
num_dense_layers: Number of dense layers.
num_dense_nodes: Number of nodes in each dense layer.
activation: Activation function for all layers.
"""
# Start construction of a Keras Sequential model.
model = Sequential()
# First convolutional layer.
# There are many hyper-parameters in this layer
# For this demo, we will optimize the activation function only.
model.add(Conv2D(kernel_size=5, strides=1, filters=16, padding='same',
activation=activation, name='layer_conv1'))
model.add(MaxPool2D(pool_size=2, strides=2))
# Second convolutional layer.
# Again, we will only optimize the activation function.
model.add(Conv2D(kernel_size=5, strides=1, filters=36, padding='same',
activation=activation, name='layer_conv2'))
model.add(MaxPool2D(pool_size=2, strides=2))
# Flatten the 4-rank output of the convolutional layers
# to 2-rank that can be input to a fully-connected Dense layer.
model.add(Flatten())
# Add fully-connected Dense layers.
# The number of layers is a hyper-parameter we want to optimize.
# We add the different number of layers in the following loop:
for i in range(num_dense_layers):
# Add the dense fully-connected layer to the model.
# This has two hyper-parameters we want to optimize:
# The number of nodes (neurons) and the activation function.
model.add(Dense(num_dense_nodes,
activation=activation,
))
# Last fully-connected dense layer with softmax-activation
# for use in classification.
model.add(Dense(10, activation='softmax'))
# Use the Adam method for training the network.
# We want to find the best learning-rate for the Adam method.
optimizer = Adam(learning_rate=learning_rate)
# In Keras we need to compile the model so it can be trained.
model.compile(optimizer=optimizer,
loss='categorical_crossentropy',
metrics=['accuracy'])
return model
# We define the hyperparameter space
# min, max, prior: distribution "uniform" (default) or "log-uniform", name
dim_learning_rate = Real(
low=1e-6,
high=1e-2,
prior='log-uniform', # sample it from teh log-uniform distribution
name='learning_rate',
)
# uniformly sampled by default
dim_num_dense_layers = Integer(low=1,
high=5,
name='num_dense_layers')
dim_num_dense_nodes = Integer(low=5, high=512, name='num_dense_nodes')
dim_activation = Categorical(
categories=['relu', 'sigmoid'], name='activation',
)
# the hyperparameter space grid
param_grid = [dim_learning_rate,
dim_num_dense_layers,
dim_num_dense_nodes,
dim_activation]
# NOTE: other possible hyperparameters: batch size, epochs, number of convolutional layers in each conv-pool block, etc.
# we will save the model with this name
path_best_model = 'cnn_model.h5'
# starting point for the optimization
best_accuracy = 0
# The objective function in the case of a neural network does the following:
# - Instantiate a model with a set of hyperparameters: create_cnn()
# - Instantiate anything required by the model, e.g., learning rate reduction scheme
# - Fit/train it for some epoches and a given fraction of validation split
# - If the score is the best so far, save the model and its training statistics
@use_named_args(param_grid)
def objective(
learning_rate,
num_dense_layers,
num_dense_nodes,
activation,
):
"""
Hyper-parameters:
learning_rate: Learning-rate for the optimizer.
num_dense_layers: Number of dense layers.
num_dense_nodes: Number of nodes in each dense layer.
activation: Activation function for all layers.
"""
# Print the hyper-parameters.
print('learning rate: {0:.1e}'.format(learning_rate))
print('num_dense_layers:', num_dense_layers)
print('num_dense_nodes:', num_dense_nodes)
print('activation:', activation)
print()
# Create the neural network with the hyper-parameters.
# We call the function we created previously.
model = create_cnn(learning_rate=learning_rate,
num_dense_layers=num_dense_layers,
num_dense_nodes=num_dense_nodes,
activation=activation)
# Set a learning rate annealer
# this reduces the learning rate if learning does not improve
# for a certain number of epochs
learning_rate_reduction = ReduceLROnPlateau(monitor='val_accuracy',
patience=2,
verbose=1,
factor=0.5,
min_lr=0.00001)
# train the model
# we use 3 epochs to be able to run the notebook in a "reasonable"
# time. If we increase the epochs, we will have better performance
# this could be another parameter to optimize in fact.
history = model.fit(x=X_train,
y=y_train,
epochs=3, # 3
batch_size=128, # this could be a hyperparameter, too
validation_split=0.1,
callbacks=learning_rate_reduction)
# Get the classification accuracy on the validation-set
# after the last training-epoch.
accuracy = history.history['val_accuracy'][-1]
# Print the classification accuracy.
print()
print("Accuracy: {0:.2%}".format(accuracy))
print()
# Save the model if it improves on the best-found performance.
# We use the global keyword so we update the variable outside
# of this function.
global best_accuracy
# If the classification accuracy of the saved model is improved ...
if accuracy > best_accuracy:
# Save the new model to harddisk.
# Training CNNs is costly, so we want to avoid having to re-train
# the network with the best found parameters. We save it instead
# as we search for the best hyperparam space.
model.save(path_best_model)
# Update the classification accuracy.
best_accuracy = accuracy
# VERY IMPORTANT!
# Delete the Keras model with these hyper-parameters from memory.
del model
# Remember that Scikit-optimize always minimizes the objective
# function, so we need to negate the accuracy (because we want
# the maximum accuracy)
return -accuracy
# Before we run the hyper-parameter optimization,
# let's first check that the everything is working
# by passing some default hyper-parameters.
# Beware of the time needed for one evaluation and
# modify epochs and n_calls accordingly
default_parameters = [1e-5, 1, 16, 'relu']
objective(x=default_parameters)
# gp_minimize performs by default GP Optimization
# using a Marten Kernel
# NOTE: instead of passing n_initial_points, we pass
# th einitial parameters!
gp_ = gp_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
x0=default_parameters, # the initial parameters to test
acq_func='EI', # the acquisition function
n_calls=30, # the number of subsequent evaluations of f(x)
random_state=0,
)
# function value at the minimum.
# note that it is the negative of the accuracy
"Best score=%.4f" % gp_.fun
# 'Best score=-0.9849'
# we can access the hyperparameter space
gp_.space
print("""Best parameters:
=========================
- learning rate=%.6f
- num_dense_layers=%d
- num_nodes=%d
- activation = %s""" % (gp_.x[0],
gp_.x[1],
gp_.x[2],
gp_.x[3]))
# Best parameters:
# =========================
# - learning rate=0.005901
# - num_dense_layers=1
# - num_nodes=512
# - activation = relu
# Always plot the convergence to check whether we founf the minimum
plot_convergence(gp_)
# Objective plot:
# We select the hyperparameters we want to analyze
# A matrix of plots is shown
# The diagonal shows the effect of each selected hyperparam on the response
# The non-diagonal plots are bi-variate plots of pairs of hyperparams
# and they also show the effect of varying hyperparam values on the response.
# In black, the sampled combinations, in red, the optimum combination (minimum response in objective)
# We see that sometimes some hyperparams have no effect, e.g., number of dense layers
# https://scikit-optimize.github.io/stable/modules/generated/skopt.plots.plot_objective.html#skopt.plots.plot_objective
dim_names = ['learning_rate', 'num_dense_nodes', 'num_dense_layers']
plot_objective(result=gp_, plot_dims=dim_names)
plt.show()
# Evaluations plot
# We select the hyperparameters we want to analyze
# A matrix of plots is shown
# The diagonal shows the histogram of samplings for each selected hyperparam
# The non-diagonal plots show the order of the sampling with pairs of hyperparams
# The order in which hyperparams were sampled is color-coded
# The optimum point is in red
# https://scikit-optimize.github.io/stable/modules/generated/skopt.plots.plot_evaluations.html
plot_evaluations(result=gp_, plot_dims=dim_names)
plt.show()
# Model Evaluation
# We need to evaluate the final/best model with the test set
# For that, we load the best model
model = load_model(path_best_model)
# make predictions in test set
# NOTE: there is actually another dataset we could use...
# but we use the split we created.
result = model.evaluate(x=X_test,
y=y_test)
# print evaluation metrics
for name, value in zip(model.metrics_names, result):
print(name, value)
# loss 0.049428313970565796
# accuracy 0.9840475916862488
# Predict the values from the validation dataset
y_pred = model.predict(X_test)
# Convert predictions classes to one hot vectors
y_pred_classes = np.argmax(y_pred, axis = 1)
# Convert validation observations to one hot vectors
y_true = np.argmax(y_test, axis = 1)
# compute the confusion matrix
cm = confusion_matrix(y_true, y_pred_classes)
cm
# Confusion matrix: let's make it more colourful
classes = 10
plt.imshow(cm, interpolation='nearest', cmap=plt.cm.Blues)
plt.title('Confusion matrix')
plt.colorbar()
tick_marks = np.arange(classes)
plt.xticks(tick_marks, range(classes), rotation=45)
plt.yticks(tick_marks, range(classes))
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, cm[i, j],
horizontalalignment="center",
color="white" if cm[i, j] > 100 else "black",
)
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')Bayesian optimization is one type of Sequential Model-Based Optimization (SMBO) method to optimize black-box functions. It can be used any time the f(x) function to optimize is very difficult/costly to evaluate, and we don't know its shape (formula) nor its derivative.
In particular, when we apply Bayesian optimization, Gaussian Processes are regressed as a surrogate of the real f(x) with observations and used to estimate the minimum/maximum point. The regression is done sequentially and the x values to be used at each step are given by the acquisition functions.
However, there are other SMBO methods different to the introduced Bayesian optimization with Gaussian Processes. These alternative methods, follow a similar approach, they are a Bayesian optimization, but the surrogate function and its regression are different. Methods introduced in this section:
- SMACs: Instead of Gaussian Processes, we can use Random Forests and Gradient Boosted Models (GBM) as the surrogate function.
- TPE: Tree-structured Parzen estimators: Non-standard Bayesian optimization.
In SMACs, instead of regressing a Gaussian Process surrogate, we regress a random forest of a gradient boosted model. Similarly, our surrogate is able to predict the values of f as mean and spread: if the tree is trained, for a given x, we navigate in the nodes to the corresponding leave node and take the mean and the standard deviation in that node. Thus, with a mean and a spread, we can apply the acquisition functions introduced so far; whereby, some other modified versions are also used.
Notebook: 01-SMAC-Scikit-Optimize-CNN.ipynb. We optimine a Keras-CNN to classify MNIST hand-written digits.
The notebook is very similar to the previous one 07-Bayesian-Optimization-CNN.ipynb, but forest_minimize() is used to optimize the objective() instead of the gp_minimize():
# we approximate f(x) using Random Forests, we could
# also approximate it with gradient boosting machines
# using gbrt_minimize instead.
# https://scikit-optimize.github.io/stable/modules/generated/skopt.forest_minimize.html#skopt.forest_minimize
# https://scikit-optimize.github.io/stable/modules/generated/skopt.gbrt_minimize.html#skopt.gbrt_minimize
# The call here is identical to the call with gp_minimize
fm_ = forest_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
x0=default_parameters, # the initial parameters to test
acq_func='EI', # the acquisition function
n_calls=30, # the number of subsequent evaluations of f(x)
random_state=0,
)Tree-structured Parzen estimators do not approximate the f(x) response function, but the distirbution of the hyperparameters x:
- Many SMBO methods estimate
P(f(x)|x) - TPE is also a SMBO method, but it estimates
P(x|f(x))
From that distribution, they reach the hyperparameter set that leads to the optium response value.
Process: we repeat this for each hyperparameter:
- The hyperparameter space is sampled and
f(x)computed. - The sampled
xvalues are classified as- Small: those which lead to a small
f(x) - Large: the rest
- The threshold is usually a quantile fraction, e.g.,
gamma = 0.25.
- Small: those which lead to a small
- For each subset, the distribution density function is computed using kernels; we basically put Gaussians in each point. The used kernels are Parzen windows. An important parameter is the spread we use for them, aka., the window size; but usually, the default value is left.
L(x): distribution of Small subsetG(x): distribution of Large subset
- Then, we draw samples from
L(x); the number of samples is also a parameter, but the default is used. For each sample, we evaluate the acquisition function, which determines which of the drawn samples has the highes likelihood of leading to a smallerf(x).- The acquisition function is composed by
LandG:(gamma + (G(x)/L(x))*(1-gamma))^(-1)
- The acquisition function is composed by
- Then, the distributions
L(x)andG(x)are updated. - We continue until we converge.
The TPE method is implemented in Hyperopt. We can modify the parameters mentioned in the process, but often the default are left.
The reason TPE is a tree-based method is that we can run it with nested hyperparameter combinations in which we even define different models (linear, trees, neural networks, etc.). We can do that because each hyperparameter is analyzed independently from the others. If we don't have the case in which we want to use a nested approach, TPEs can lead to worse performance, because the inter-relationships between hyperparameters are ignored; but if we need to test nested hyperparameter sets, TPE is the choice. Another method which allows for nested testing is the good old random search.
The notebook where the default TPE is tried with a Keras-CNN is very similar to the previous one, but the Hyperopt library is used: 03-TPE-with-CNN.ipynb.
Hyperopt needs also the definition of the objective() function, and optionally, the object Trials() to record the data from the calls; then, the objective() is passed to the minimization function fmin(). That optimization API also accepts the algorithm which will be used, i.e., tpe.suggest. We can use other minimization algorithms, such as simulated annealing, which is passed as anneal.suggest.
Interesting links:
# For reproducible results.
# See:
# https://keras.io/getting_started/faq/#how-can-i-obtain-reproducible-results-using-keras-during-development
import os
os.environ['PYTHONHASHSEED'] = '0'
import numpy as np
import tensorflow as tf
import random as python_random
# The below is necessary for starting Numpy generated random numbers
# in a well-defined initial state.
np.random.seed(123)
# The below is necessary for starting core Python generated random numbers
# in a well-defined state.
python_random.seed(123)
# The below set_seed() will make random number generation
# in the TensorFlow backend have a well-defined initial state.
# For further details, see:
# https://www.tensorflow.org/api_docs/python/tf/random/set_seed
tf.random.set_seed(1234)
import itertools
from functools import partial
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from keras.utils.np_utils import to_categorical
from keras.models import Sequential, load_model
from keras.layers import Dense, Flatten, Conv2D, MaxPool2D
from keras.optimizers import Adam
from keras.callbacks import ReduceLROnPlateau
from hyperopt import hp, tpe, fmin, Trials
# Load the data
data = pd.read_csv("../data/train.csv")
# first column is the target, the rest of the columns
# are the pixels of the image
# each row is 1 image
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
data.drop(['label'], axis=1), # the images
data['label'], # the target
test_size = 0.1,
random_state=0)
# Re-scale the data
# 255 is the maximum value a pixel can take
X_train = X_train / 255
X_test = X_test / 255
# Reshape image in 3 dimensions:
# height: 28px X width: 28px X channel: 1
X_train = X_train.values.reshape(-1,28,28,1)
X_test = X_test.values.reshape(-1,28,28,1)
# the target is 1 variable with the 9 different digits
# as values
y_train.unique()
# 2, 0, 7, 4, 3, 5, 9, 6, 8, 1
# For Keras, we need to create 10 dummy variables,
# one for each digit
# Encode labels to one hot vectors (ex : digit 2 -> [0,0,1,0,0,0,0,0,0,0])
y_train = to_categorical(y_train, num_classes = 10)
y_test = to_categorical(y_test, num_classes = 10)
# Some image examples
g = plt.imshow(X_train[0][:,:,0])
# determine the hyperparameter space
# http://hyperopt.github.io/hyperopt/getting-started/search_spaces/
# NOTE: q-distributions return a float, thus, sometimes recasting needs to be done!
# hp.choice: returns one of several options (suitable for categorical hyperparams)
# hp.randint: returns a random integer between 0 and an upper limit
# hp.uniform: returns a value uniformly between specified limits
# hp.quniform: Returns a value like round(uniform(low, high) / q) * q, i.e. sample between min and max in steps of size q
# hp.loguniform: draws values from exp(uniform(low, high)) so that the logarithm of the returned value is uniformly distributed
# hp.qloguniform: Returns a value like round(exp(uniform(low, high)) / q) * q (similar use and cautions to hp.quniform but for log-uniform distributions)
# hp.normal: draws from a normal distribution with specified mu and sigma
# hp.qnormal: Returns a value like round(normal(mu, sigma) / q) * q
# hp.lognormal: Returns a value drawn according to exp(normal(mu, sigma)) so that the logarithm of the return value is normally distributed
# hp.qlognormal: Returns a value like round(exp(normal(mu, sigma)) / q) * q
param_grid = {
'learning_rate': hp.uniform('learning_rate', 1e-6, 1e-2),
'num_conv_layers': hp.quniform('num_conv_layers', 1, 3, 1),
'num_dense_layers': hp.quniform('num_dense_layers', 1, 5, 1),
'num_dense_nodes': hp.quniform('num_dense_nodes', 5, 512, 1),
'activation': hp.choice('activation', ['relu', 'sigmoid']),
}
# function to create the CNN
def create_cnn(
# the hyperparam to optimize are passed
# as arguments
learning_rate,
num_conv_layers,
num_dense_layers,
num_dense_nodes,
activation,
):
"""
Hyper-parameters:
learning_rate: Learning-rate for the optimizer.
convolutional layers: Number of conv layers.
num_dense_layers: Number of dense layers.
num_dense_nodes: Number of nodes in each dense layer.
activation: Activation function for all layers.
"""
# Start construction of a Keras Sequential model.
model = Sequential()
# First convolutional layer.
# There are many hyper-parameters in this layer
# For this demo, we will optimize the activation function and
# the number of convolutional layers that it can take.
# We add the different number of conv layers in the following loop:
for i in range(num_conv_layers):
model.add(Conv2D(kernel_size=5, strides=1, filters=16, padding='same',
activation=activation))
model.add(MaxPool2D(pool_size=2, strides=2))
# Second convolutional layer.
# Same hyperparameters to optimize as previous layer.
for i in range(num_conv_layers):
model.add(Conv2D(kernel_size=5, strides=1, filters=36, padding='same',
activation=activation))
model.add(MaxPool2D(pool_size=2, strides=2))
# Flatten the 4-rank output of the convolutional layers
# to 2-rank that can be input to a fully-connected Dense layer.
model.add(Flatten())
# Add fully-connected Dense layers.
# The number of layers is a hyper-parameter we want to optimize.
# We add the different number of layers in the following loop:
for i in range(num_dense_layers):
# Add the dense fully-connected layer to the model.
# This has two hyper-parameters we want to optimize:
# The number of nodes (neurons) and the activation function.
model.add(Dense(num_dense_nodes,
activation=activation,
))
# Last fully-connected dense layer with softmax-activation
# for use in classification.
model.add(Dense(10, activation='softmax'))
# Use the Adam method for training the network.
# We want to find the best learning-rate for the Adam method.
optimizer = Adam(lr=learning_rate)
# In Keras we need to compile the model so it can be trained.
model.compile(optimizer=optimizer,
loss='categorical_crossentropy',
metrics=['accuracy'])
return model
# we will save the model with this name
path_best_model = 'cnn_model.h5'
# starting point for the optimization
best_accuracy = 0
def objective(params):
"""
Hyper-parameters:
learning_rate: Learning-rate for the optimizer.
convolutional layers: Number of conv layers.
num_dense_layers: Number of dense layers.
num_dense_nodes: Number of nodes in each dense layer.
activation: Activation function for all layers.
"""
# Print the hyper-parameters.
print('learning rate: ', params['learning_rate'])
print('num_conv_layers: ', int(params['num_conv_layers']))
print('num_dense_layers: ',int(params['num_dense_layers']))
print('num_dense_nodes: ', int(params['num_dense_nodes']))
print('activation: ', params['activation'])
print()
# Create the neural network with the hyper-parameters.
# We call the function we created previously.
model = create_cnn(learning_rate=params['learning_rate'],
num_conv_layers=int(params['num_conv_layers']),
num_dense_layers=int(params['num_dense_layers']),
num_dense_nodes=int(params['num_dense_nodes']),
activation=params['activation'],
)
# Set a learning rate annealer
# this reduces the learning rate if learning does not improve
# for a certain number of epochs
learning_rate_reduction = ReduceLROnPlateau(monitor='val_accuracy',
patience=2,
verbose=1,
factor=0.5,
min_lr=0.00001)
# train the model
# we use 3 epochs to be able to run the notebook in a "reasonable"
# time. If we increase the epochs, we will have better performance
# this could be another parameter to optimize in fact.
history = model.fit(x=X_train,
y=y_train,
epochs=3,
batch_size=128,
validation_split=0.1,
callbacks=learning_rate_reduction,
verbose=0) # on Windows, all steps of the progress bar are saved...
# Get the classification accuracy on the validation-set
# after the last training-epoch.
accuracy = history.history['val_accuracy'][-1]
# Print the classification accuracy.
print()
print("Accuracy: {0:.2%}".format(accuracy))
print()
# Save the model if it improves on the best-found performance.
# We use the global keyword so we update the variable outside
# of this function.
global best_accuracy
# If the classification accuracy of the saved model is improved ...
if accuracy > best_accuracy:
# Save the new model to harddisk.
# Training CNNs is costly, so we want to avoid having to re-train
# the network with the best found parameters. We save it instead
# as we search for the best hyperparam space.
model.save(path_best_model)
# Update the classification accuracy.
best_accuracy = accuracy
# Delete the Keras model with these hyper-parameters from memory.
del model
# Remember that Scikit-optimize always minimizes the objective
# function, so we need to negate the accuracy (because we want
# the maximum accuracy)
return -accuracy
# Before we run the hyper-parameter optimization,
# let's first check that the everything is working
# by passing some default hyper-parameters.
default_parameters = {
'learning_rate': 1e-5,
'num_conv_layers': 1,
'num_dense_layers': 1,
'num_dense_nodes': 16,
'activation': 'relu',
}
# Test objective function
objective(default_parameters)
# learning rate: 1e-05
# num_conv_layers: 1
# num_dense_layers: 1
# num_dense_nodes: 16
# activation: relu
# Accuracy: 51.69%
# fmin performs the minimization
# tpe.suggest samples the parameters following tpe
# SMBO: TPE
# fmin performs the minimization
# tpe.suggest samples the parameters following tpe
# with default parameters for TPE.
# We can pass another algorithm, too, e.g.: anneal.suggest
# this one is the simulated annealing algorithm:
# https://en.wikipedia.org/wiki/Simulated_annealing
# https://www.kaggle.com/code/ilialar/hyperparameters-tunning-with-hyperopt
trials = Trials()
search = fmin(
fn=objective,
space=param_grid,
max_evals=10, #30,
rstate=np.random.default_rng(42),
algo=tpe.suggest, # tpe
#algo=anneal.suggest, # simulated annealing
trials=trials
)
# tpe.suggest default parameters:
# https://github.com/hyperopt/hyperopt/blob/master/hyperopt/tpe.py#L781
# to override the defo parameters:
# https://github.com/hyperopt/hyperopt/issues/632
# the best hyperparameters can also be found in
# trials
trials.argmin
results = pd.concat([
pd.DataFrame(trials.vals),
pd.DataFrame(trials.results)],
axis=1,
).sort_values(by='loss', ascending=False).reset_index(drop=True)
results['loss'].plot()
plt.ylabel('Accuracy')
plt.xlabel('Hyperparam combination')Comparison between tuning algorithms
Comparison between tuning algorithms
Summary:
- For smaller models use
RandomSearchCV. - For bigger models, staring with trees, try also
BayesSearchCV. - For biggest models (neural networks), try SMAC, Bayes Search or TPE.
In this section, methods that use different types of fidelities of f(x) qualities are briefly introduced.
As seen until now:
- Random search is the best method for small-medium-sized models; but it is computation intensive.
- Sequential model-based optimization methods (Bayesian optimization, SMACs, TPE) are better suited for medium-large models in which evaluating the
f(x)response surface is expensive; but these methods are time intensive.
Multi-fidelity optimization methods achieve good results on a time budget; they do that evaluating a cheaper version of f(x); in fact they combine the results of:
- Many cheap
f(x)evaluations - Few accurate
f(x)evaluations
A cheap f(x) evaluation/representation can be achieved with:
- Reduced dataset
- Reduced number of features
- Early stopping
- Cheaper value of a hyperparameter
One implementation of a multi-fidelity method is in Scikit-Learn: Successive Halving: HalvingGridSearchCV, HalvingRandomSearchCV.
This section is empty. I leave it here so that the notebook folder names coincide with the sections in this guide. It was probably a bug.
This section is a review/summary of the Scikit-Optimize library. All the theoretical concepts were already introduced, as well as almost all skopt APIs.
There are two main APIs for Bayesian optimization in skopt:
- The manual API, in which we define the
objective()function which performs the model training given a set of hyperparameters. Here, we can use any model and any cross-validation scheme. For instance, we can use scikit-learn models withcross_val_score(). Theobjective()function is passed to a search algorithm, e.g.,gp_minimize(). The manual API is more complex, but provides a lot of flexibility, so that we can optimize any model of any ML framework with it! - The automatic API for Bayesian optimization, which mimicks the Scikit-Learn objects: we instantiate
BayesSearchCVand pass to it the hyperparameter space and asklearn-conform estimator; then, everything is done automatically, as it were aGridSearchCVobject.
Notes:
- Scikit-optimize is very sensitive to the Scikit-Learn and Scipy versions used; additionally, it seems there are no new versions of Scikit-Optimize in the past 2 years. Sometimes some examples from the course yield errors, probably due to version conflicts. Used versions:
numpy==1.22.0 scipy==1.5.3 scikit-learn==0.23.2 scikit-optimize==0.8.1 - Maybe an important remark is that Scikit-Optimize is currently not updated/maintained regularly; the last release to date (2023-05) is v0.9 in 2021-10.
- Always plot the convergence with
plot_convergence(). We can see how many iterations are really necessary; even though we might usen_calls=50often times, sometimes30calls is more than enough. - To analyze the optimization process (i.e., response as function of hyperparameters and hyperparameter values in time), use, respectively:
plot_objective()andplot_evaluations().
For a more detailed information, check these sections, with their respective notebooks:
- Section 5 - Random Search with Scikit-Optimize
- Section 6 - Scikit-Optimize for Bayesian Optimization: Notes
- Section 6 - Example: Automatic Gaussian Optimization of a Grandient Boosted Tree with 4 Hyperparameters (BayesSearchCV)
- Section 6 - Example: Manual Bayesian Optimization of an XGBoost Classifier
- Section 6 - Example: Manual Bayesian Optimization of a Keras-CNN
- Section 7 - SMACs: Sequential Model-Based Algorithm Configuration: Using Tree-Based Models as Surrogates
Hyperparameter space definition is done in a list which takes distributions with built-in objects:
- Samples Reals, Integers and Categories
- For Reals and Integers samples with uniform and log-uniform
# In the manual API, the hyperparameter space is a list
# In the automatic/sklearn API the hyperparameter space is a dictionary
# min, max, prior: distribution "uniform" (default) or "log-uniform", name
param_grid = [
Integer(10, 120, name="n_estimators"), # prior is "uniform" by default
Integer(1, 5, name="max_depth"),
Real(0.0001, 0.1, prior='log-uniform', name='learning_rate'),
Real(0.001, 0.999, prior='log-uniform', name="min_samples_split"),
Categorical(['deviance', 'exponential'], name="loss"),
]
# to sample a parameter value, i.e., draw a random value from its distribution
param_grid.rvs()[0]A typical objective function looks like this:
# instantiate model
# NOTE: we can either
# 1. instantiate the model outside and then model.set_params(**params)
# 2. or instantiate a new model inside objective() (as done in the example with the CNN)
# In the case of a CNN, we `del model` at the end in objective() to free memory
# I would argue that it's cleaner to instantiate everything inside objective()
model = GradientBoostingClassifier(random_state=0)
# we pass the hyperparameter space via a decorator
@use_named_args(param_grid)
def objective(**params):
model.set_params(**params)
value = np.mean(
# cross-validation from sklearn
cross_val_score(
model,
X_train,
y_train,
cv=3,
n_jobs=-4,
scoring='accuracy')
)
# negate because we need to minimize
return -valueAvailable search algorithms that take the objective and the hyperparameter space:
gp_minimize: Bayesian optimization with Gaussian Process Regression.forest_minimize: SMAC: Bayesian optimization with Random Forests.gbrt_minimize: Bayesian Optimization with Gradient Boosted Trees.dummy_minimize: Random search.
# Bayesian optimization with Gaussian Process Regression
gp_ = gp_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
n_initial_points=10, # the number of points to evaluate f(x) to start of
acq_func='EI', # the acquisition function
n_calls=30, # the number of subsequent evaluations of f(x)
random_state=0,
)
# SMAC: Bayesian optimization with Random Forests
fm_ = forest_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
base_estimator = 'RF', # the surrogate
n_initial_points=10, # the number of points to evaluate f(x) to start of
acq_func='EI', # the acquisition function
n_calls=30, # the number of subsequent evaluations of f(x)
random_state=0,
n_jobs=4,
)
# Bayesian Optimization with Gradient Boosted Trees
gbm_ = gbrt_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
n_initial_points=10, # the number of points to evaluate f(x) to start of
acq_func='EI', # the acquisition function
n_calls=30, # the number of subsequent evaluations of f(x)
random_state=0,
n_jobs=4,
)
# Random search
search = dummy_minimize(
objective, # the objective function to minimize
param_grid, # the hyperparameter space
n_calls=50, # the number of evaluations of the objective function
random_state=0,
)Built-in acquisition functions passed to the search/optimization function via acq_func:
- Expected Improvement (EI):
"EI" - Probability of Improvement (PI):
"PI" - Lower Confidence Bound (LCB):
"LCB" - EI and PI per second, to account for compute time:
"EIps","PIps" - Hedge optimization of all acquisition functions:
"gp_hedge"
Results and analysis of the optimization:
# The following calls are for the manual API
# The object returned by the search/optimization has many information!
search
# Best score
search.fun
# Best hyperparameters (index is definition order in hyperparameter space/list)
search.x[0]
search.x[1]
# We can see how many iterations are really necessary, and whether we reached the optimum
plot_convergence(search)
# It shows the selection of hyperparameter values along the different calls
params_examine = ['param_name_1', 'param_name_2']
plot_evaluations(search, plot_dims=params_examine)
# It shows the response as function of different hyperparameter values
params_examine = ['param_name_1', 'param_name_2']
plot_objective(search, plot_dims=params_examine)
# Create a dataframe with all trials
experiment_stats = pd.concat([
pd.DataFrame(search.x_iters),
pd.Series(search.func_vals),
], axis=1)
experiment_stats.columns = params_examine + ['accuracy'] # or the score selected in objective
experiment_stats.sort_values(by='accuracy', ascending=True, inplace=True)
experiment_stats.head()This is the list of the example notebooks in this section:
01-Search-space-distributions.ipynb02-Randomized-Search.ipynb03-Bayesian-Optimization-GP-Manual.ipynb04-Bayesian-Optimization-Random-Forest.ipynb05-Bayesian-Optimization-GBM.ipynb06-Parallelization.ipynb07-Bayesian-Optimization-Sklearn-wrapper.ipynb08-Bayesian-Optimization-Change-Kernel.ipynb09-Bayesian-Optimization-xgb-Optional.ipynb10-Bayesian-Optimization-CNN.ipynb
These example notebooks in this section do not introduce almost anything new since the content in most of them has been implemented in prior sections; the exception is the parallelization notebook, which shows how we can parallelize search/optimization algorithms. Recall that random and grid searches are easily parallelizable because each trial is independent from the other; in contrast, sequential methods (Bayesian search with Gaussian processes, SMACs, etc.) are difficult to parallelize, because each trial depends on the previous. The way they are parallelized is with trial branches: each thread/sub-process works on a sequence of trials.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import cross_val_score, train_test_split
from skopt import Optimizer # for the optimization
from joblib import Parallel, delayed # for the parallelization
from skopt.space import Real, Integer, Categorical
from skopt.utils import use_named_args
# load dataset
breast_cancer_X, breast_cancer_y = load_breast_cancer(return_X_y=True)
X = pd.DataFrame(breast_cancer_X)
y = pd.Series(breast_cancer_y).map({0:1, 1:0})
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0)
# Scikit-optimize parameter grid is a list
param_grid = [
Integer(10, 120, name="n_estimators"),
Integer(1, 5, name="max_depth"),
Real(0.0001, 0.1, prior='log-uniform', name='learning_rate'),
Real(0.001, 0.999, prior='log-uniform', name="min_samples_split"),
Categorical(['deviance', 'exponential'], name="loss"),
]
# set up the gradient boosting classifier
gbm = GradientBoostingClassifier(random_state=0)
# We design a function to maximize the accuracy, of a GBM,
# with cross-validation
# the decorator allows our objective function to receive the parameters as
# keyword arguments. This is a requirement for scikit-optimize.
@use_named_args(param_grid)
def objective(**params):
# model with new parameters
gbm.set_params(**params)
# optimization function (hyperparam response function)
value = np.mean(
cross_val_score(
gbm,
X_train,
y_train,
cv=3,
n_jobs=-4,
scoring='accuracy')
)
# negate because we need to minimize
return -value
# We use the Optimizer
# https://scikit-optimize.github.io/stable/modules/generated/skopt.Optimizer.html
# In contrast to other optimization functions,
# this one doesn't run the entire optimization when instantiated,
# instead, we need to run optimizer.ask() on it
# which we do in different threads using joblib.Parallel.
# We define the number of parallel jobs with
# n_jobs=n_points = 4 parallel jobs
# Additionally, the number of initial evaluations is defined here, too
# n_initial_points=10
n_points = 4
optimizer = Optimizer(
dimensions = param_grid, # the hyperparameter space
base_estimator = "GP", # the surrogate; options: "GP", "RF", "ET", "GBRT"
n_initial_points=10, # the number of points to evaluate f(x) to start of
acq_func='EI', # the acquisition function
random_state=0,
n_jobs=n_points,
)
# Now, we run the calls in parallel,
# we will use 4 CPUs, i.e., 4 parallel jobs (n_points);
# if we loop 10 times using 4 end points, we perform 40 searches in total
n_calls = 10
for i in range(n_calls):
# n_points: number of parallel jobs
x = optimizer.ask(n_points=n_points) # x is a list of parallel hyperparameter spaces
y = Parallel(n_jobs=n_points)(delayed(objective)(v) for v in x) # evaluate points in parallel
optimizer.tell(x, y)
# the evaluated hyperparamters: X
optimizer.Xi
# the accuracy: y, the selected score in the objective function
optimizer.yi
# all together in one dataframe, so we can investigate further
dim_names = ['n_estimators', 'max_depth', 'min_samples_split', 'learning_rate', 'loss']
tmp = pd.concat([
pd.DataFrame(optimizer.Xi),
pd.Series(optimizer.yi),
], axis=1)
tmp.columns = dim_names + ['accuracy']
tmp.head()
# This is equivalent to plot_convergence()
tmp['accuracy'].sort_values(ascending=False).reset_index(drop=True).plot()This section is a review/summary of the Hyperopt library. All the theoretical concepts were already introduced, as well as some hyperopt APIs.
Notes:
- The library is not maintained that frequently. The last release tag v0.2.7 as of today (2023-05) is from 2021-11.
- The documentation is not very extensive.
- Hyperopt has a Scikit-Learn-specific library hyperopt-sklearn, which targets
sklearnobjects specifically; however, it doesn't seem to have a widespread adoption.
List of past sections/notebooks:
- Section 5 - Random Search with Hyperopt
- Section 7 - TPE: Tree-Structured Parzen Estimators with Hyperopt
The advantages compared to Scikit-Optimize could be:
- Implementation of TPE.
- Advanced hyperparameter spaces: many distributions and nested spaces.
Hyperparameter space definition is done in a dictionary which takes distributions with built-in objects from the hp module:
- Samples Reals, Integers and Categories.
- In contrast to Scikit-Optimize, we can sample from many distributions.
- It accepts also nested hyperparameter spaces, i.e., hyperparameters that depend on other hyperparameters; this is the major advantage of Hyperopt. We can even try different models in the tuning process.
# http://hyperopt.github.io/hyperopt/getting-started/search_spaces/
# NOTE: q-distributions return a float, thus, sometimes recasting needs to be done!
# hp.choice: returns one of several options (suitable for categorical hyperparams)
# hp.randint: returns a random integer between 0 and an upper limit
# hp.uniform: returns a value uniformly between specified limits
# hp.quniform: Returns a value like round(uniform(low, high) / q) * q, i.e. sample between min and max in steps of size q
# hp.loguniform: draws values from exp(uniform(low, high)) so that the logarithm of the returned value is uniformly distributed
# hp.qloguniform: Returns a value like round(exp(uniform(low, high)) / q) * q (similar use and cautions to hp.quniform but for log-uniform distributions)
# hp.normal: draws from a normal distribution with specified mu and sigma
# hp.qnormal: Returns a value like round(normal(mu, sigma) / q) * q
# hp.lognormal: Returns a value drawn according to exp(normal(mu, sigma)) so that the logarithm of the return value is normally distributed
# hp.qlognormal: Returns a value like round(exp(normal(mu, sigma)) / q) * q
param_grid = {
'n_estimators': hp.quniform('n_estimators', 200, 2500, 100), # min, max, step
'max_depth': hp.uniform('max_depth', 1, 10), # min, max
'learning_rate': hp.uniform('learning_rate', 0.01, 0.99),
'booster': hp.choice('booster', ['gbtree', 'dart']),
'gamma': hp.quniform('gamma', 0.01, 10, 0.1),
'subsample': hp.uniform('subsample', 0.50, 0.90),
'colsample_bytree': hp.uniform('colsample_bytree', 0.50, 0.99),
'colsample_bylevel': hp.uniform('colsample_bylevel', 0.50, 0.99),
'colsample_bynode': hp.uniform('colsample_bynode', 0.50, 0.99),
'reg_lambda': hp.uniform('reg_lambda', 1, 20)
}
# Example of a nested hyperparameter space
space = hp.choice('classifier_type', [
{
'type': 'naive_bayes',
},
{
'type': 'svm',
'C': hp.lognormal('svm_C', 0, 1),
'kernel': hp.choice('svm_kernel', [
{'ktype': 'linear'},
{'ktype': 'RBF', 'width': hp.lognormal('svm_rbf_width', 0, 1)},
]),
},
{
'type': 'dtree',
'criterion': hp.choice('dtree_criterion', ['gini', 'entropy']),
'max_depth': hp.choice('dtree_max_depth',
[None, hp.qlognormal('dtree_max_depth_int', 3, 1, 1)]),
'min_samples_split': hp.qlognormal('dtree_min_samples_split', 2, 1, 1),
},
])
# another nested space in which the models itself is a hyperparameter,
# i.e., we try different models/algorithms
param_grid = hp.choice('classifier', [
# algo 1
{'model': LogisticRegression,
'params': {
'penalty': hp.choice('penalty', ['l1','l2']),
'C' : hp.uniform('C', 0.001, 10),
'solver': 'saga', # the only solver that works with both penalties
}},
# algo 2
{'model': RandomForestClassifier,
'params': {
'n_estimators': hp.quniform('n_estimators_rf', 50, 1500, 50),
'max_depth': hp.quniform('max_depth_rf', 1, 5, 1),
'criterion': hp.choice('criterion_rf', ['gini', 'entropy']),
}},
# algo 3
{'model': GradientBoostingClassifier,
'params': {
'n_estimators': hp.quniform('n_estimators_gbm', 50, 1500, 50),
'max_depth': hp.quniform('max_depth_gbm', 1, 5, 1),
'criterion': hp.choice('criterion_gbm', ['friedman_mse', 'mse']),
}},
])A typical objective function looks like this:
# the objective function takes one instance from
# the hyperparameter space as input, not the complete param_grid
def objective(params):
# we need a dictionary to indicate which value from the space
# to attribute to each value of the hyperparameter in the xgb
params_dict = {
# important int, as it takes integers only
'n_estimators': int(params['n_estimators']),
# important int, as it takes integers only
'max_depth': int(params['max_depth']),
'learning_rate': params['learning_rate'],
'booster': params['booster'],
'gamma': params['gamma'],
'subsample': params['subsample'],
'colsample_bytree': params['colsample_bytree'],
'colsample_bylevel': params['colsample_bylevel'],
'colsample_bynode': params['colsample_bynode'],
'random_state': 1000,
}
# As oposed to Scikit-Optimize, the model is usually instantiated
# in the objective()
# with ** we pass the items in the dictionary as parameters
# to the xgb, our model in this example
model = xgb.XGBClassifier(**params_dict)
# train with cv
score = cross_val_score(model,
X_train,
y_train,
scoring='accuracy',
cv=3,
n_jobs=4).mean()
# to minimize, we negate the score
return -scoreAvailable search algorithms passed to the optimization function fmin() via algo:
rand.suggest: Random search.anneal.suggest: Simulated Annealing, a Sequential Model-Based approach.tpe.suggest: Tree-structured Parzen estimators (TPE), another Sequential Model-Based approach.
# Track experiment metrics
trials = Trials()
# fmin performs the minimization
search = fmin(
fn=objective,
space=param_grid,
max_evals=50, # number of combinations we'd like to randomly test
rstate=np.random.default_rng(42),
algo=rand.suggest, # Randomized search
#algo=anneal.suggest`, # Simulated Annealing
#algo=tpe.suggest, # Tree-structured Parzen estimators (TPE)
trials=trials
)It has a unique acquisition function Expected Improvement (EI); it is evaluated in two ways:
- EI evaluated at binomial distributions of the input space for discrete and categorical hyperparameters
- EI evaluated with CMA-ES for the continuous hyperparameter space
Results and analysis of the optimization:
# The best parameters are in the object returned by fmin
# and also in the (optional) Trials object
search
trials.argmin
# more information from trials
trials.average_best_error() # best score
trials.best_trial # dict with infor on best trial
# convergence plot
pd.Series(trials.losses()).sort_values(ascending=False).reset_index(drop=True).plot()
# we can now extract the best hyperparameters and train a new final model
def create_param_grid(search, booster):
best_hp_dict = {
'n_estimators': int(search['n_estimators']), # important int, as it takes integers only
'max_depth': int(search['max_depth']), # important int, as it takes integers only
'learning_rate': search['learning_rate'],
'booster': booster,
'gamma': search['gamma'],
'subsample': search['subsample'],
'colsample_bytree': search['colsample_bytree'],
'colsample_bylevel': search['colsample_bylevel'],
'colsample_bynode': search['colsample_bynode'],
'random_state': 1000,
}
return best_hp_dict
best_params = create_param_grid(random_search, 'gbtree')
gbm = xgb.XGBClassifier(**best_params)
gbm.fit(X_train, y_train)
# create a dataframe with all hyperparam combinations + results
results = pd.concat([
pd.DataFrame(trials.vals),
pd.DataFrame(trials.results)],
axis=1,
).sort_values(by='loss', ascending=False).reset_index(drop=True)
# plot convergence again
results['loss'].plot()
plt.ylabel('Accuracy')
plt.xlabel('Hyperparam combination')There are also plotting functions, but they are not documented poperly: plotting.
This is the list of the example notebooks in this section:
01-Hyperparameter-Distributions.ipynb02-Search-algorithms.ipynb03-Evaluating-the-search.ipynb04-Conditonal-Search-Spaces.ipynb05-TPE-with-CNN.ipynb06-Optional-Sklearn-MLPClassifier.ipynb
This framework is new: it is described in the course, but it wasn't introudced until now.
Optuna is a very nice hyperparameter optimization engine which seems to be called to replace the previous ones. Links:
Main advantages, features:
- Pythonic, easy to use.
- Nice documentation and code examples for the most important ML libraries.
- The
objective()function created by the user, as in the other libraries, and that's where almost everything related to the tuning is specified. - We create the hyperparameter space within the
objective()using python, i.e., we can create easy-to-understand nested/conditional spaces, the model and everythin we need in theobjective(); this is called design-by-run API. For the hyperparameter space, we can use, as always, reals, integers and categoricals, and the distrubutions are currently the uniform and the log-uniform. - Many and poweful sampling algorithms:
- Grid search, random search.
- TPE: Tree-structured Parzen Estimators.
- CMA-ES
- Multi-objective samplers: NSGA-II or MOTPE.
- The acquisition function cannot be chosen, it is part of the algorithm; used functions:
- Expected Improvement (EI)
- Expected hypervolume improvement (EHVI)
- We can set a pruner, i.e., a method by which we stop the optimization if the obtained performance/score is not being good.
- Search analysis is great:
- The search/study returns a dataframe.
- We can store the results in a SQLite databse, and perform queries in parallel; we can also load the SQLite database and keep on with trials later on.
- We can do the analysis loading the SQLite database, too.
- We have many built-in functions for plotting, both with a Matplotlib and a Plotly (interactive) backend.
- There is a dashboard for the analysis.
The following overview is given using the code from the notebook
02-Conditional-Spaces-Multiple-Models.ipynb:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer
from sklearn.metrics import accuracy_score, roc_auc_score
from sklearn.model_selection import cross_val_score, train_test_split
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.linear_model import LogisticRegression
import optuna
breast_cancer_X, breast_cancer_y = load_breast_cancer(return_X_y=True)
X = pd.DataFrame(breast_cancer_X)
y = pd.Series(breast_cancer_y).map({0:1, 1:0})
# the dataset is defined outside, but used in objective()
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0)
# objective function with nested spaces
def objective(trial):
# The trial object is created under the hood, we don't instantiate it manually
# https://optuna.readthedocs.io/en/stable/reference/generated/optuna.trial.Trial.html
# We can create hyperparameter spaces with it:
# suggest_categorical(): Suggest a value for the categorical parameter.
# suggest_discrete_uniform(name, low, high, q): Suggest a value for the discrete parameter.
# suggest_float(name, low, high, *[, step, log]): Suggest a value for the floating point parameter.
# suggest_int(name, low, high[, step, log]): Suggest a value for the integer parameter.
# suggest_loguniform(name, low, high): Suggest a value for the continuous parameter.
# suggest_uniform(name, low, high): Suggest a value for the continuous parameter.
# first hypperparameter: model type
classifier_name = trial.suggest_categorical("classifier", ["LR", "RF", 'GBM'])
# depending on the model: define specific space + model
if classifier_name == "LR":
# define hyperparameter space of model
logit_penalty = trial.suggest_categorical('logit_penalty', ['l1','l2'])
logit_c = trial.suggest_float('logit_c', 0.001, 10)
logit_solver = 'saga'
# instantiate model
model = LogisticRegression(
penalty=logit_penalty,
C=logit_c,
solver=logit_solver,
)
elif classifier_name =="RF":
rf_n_estimators = trial.suggest_int("rf_n_estimators", 100, 1000)
rf_criterion = trial.suggest_categorical("rf_criterion", ['gini', 'entropy'])
rf_max_depth = trial.suggest_int("rf_max_depth", 1, 4)
rf_min_samples_split = trial.suggest_float("rf_min_samples_split", 0.01, 1)
model = RandomForestClassifier(
n_estimators=rf_n_estimators,
criterion=rf_criterion,
max_depth=rf_max_depth,
min_samples_split=rf_min_samples_split,
)
else:
gbm_n_estimators = trial.suggest_int("gbm_n_estimators", 100, 1000)
gbm_criterion = trial.suggest_categorical("gbm_criterion", ['mse', 'friedman_mse'])
gbm_max_depth = trial.suggest_int("gbm_max_depth", 1, 4)
gbm_min_samples_split = trial.suggest_float("gbm_min_samples_split", 0.01, 1)
model = GradientBoostingClassifier(
n_estimators=gbm_n_estimators,
criterion=gbm_criterion,
max_depth=gbm_max_depth,
min_samples_split=gbm_min_samples_split,
)
# cross-validation: dataset is defined outside
score = cross_val_score(model, X_train, y_train, cv=3)
accuracy = score.mean()
return accuracy
# create an optimization study: we pass the sampling/search algorithm here!
study = optuna.create_study(
# we can minimize/maximize
direction="maximize",
# we can remove this line, TPS is default
sampler=optuna.samplers.TPESampler(),
)
# Parameters of create_study()
# https://optuna.readthedocs.io/en/stable/reference/generated/optuna.study.create_study.html#optuna.study.create_study
# - storage: url to a database
# - sampler: the hyperparameter search algorithm (dafault: TPE)
# optuna.samplers.GridSampler(): in this case the param_grid is a dict with lists defined outside from objective() and passed to GridSampler() directly
# optuna.samplers.RandomSampler()
# optuna.samplers.TPESampler(): default
# optuna.samplers.CmaEsSampler()
# ...
# - pruner: the algorithm to prune unsuccessful trials (default: MedianPruner); the optimization stops if the performances are not good
# optuna.pruners.MedianPruner()
# optuna.pruners.NopPruner()
# optuna.pruners.PercentilePruner()
# ...
# - direction: 'minimize' or 'maximize'
# - study_name: name for the study, in case saved and retrieved later
# run the optimization: we pass to it the objective and the number of trials
study.optimize(objective, n_trials=20)
# best hyperparameters
study.best_params
# {'classifier': 'GBM',
# 'gbm_n_estimators': 301,
# 'gbm_criterion': 'mse',
# 'gbm_max_depth': 2,
# 'gbm_min_samples_split': 0.7450851430645121}
# best score
study.best_value
# dataframe with all trials: ech row is a trial
study.trials_dataframe()
# manual convergence plot
results['value'].sort_values().reset_index(drop=True).plot()
plt.title('Convergence plot')
plt.xlabel('Iteration')
plt.ylabel('Accuracy')This is the list of example notebooks:
01-Search-algorithms.ipynb02-Conditional-Spaces-Multiple-Models.ipynb03-Optimizing-a-CNN.ipynb04-Optimizing-a-CNN-extended.ipynb05-Evaluating-the-search.ipynb
In the following, the code from the most important notebooks is added. Note that the API Overview section already gives a very good usage example.
Notebook: 03-Optimizing-a-CNN.ipynb.
# For reproducible results.
# See:
# https://keras.io/getting_started/faq/#how-can-i-obtain-reproducible-results-using-keras-during-development
import os
os.environ['PYTHONHASHSEED'] = '0'
import numpy as np
import tensorflow as tf
import random as python_random
# The below is necessary for starting Numpy generated random numbers
# in a well-defined initial state.
np.random.seed(123)
# The below is necessary for starting core Python generated random numbers
# in a well-defined state.
python_random.seed(123)
# The below set_seed() will make random number generation
# in the TensorFlow backend have a well-defined initial state.
# For further details, see:
# https://www.tensorflow.org/api_docs/python/tf/random/set_seed
tf.random.set_seed(1234)
import itertools
from functools import partial
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from keras.utils.np_utils import to_categorical
from keras.models import Sequential, load_model
from keras.layers import Dense, Flatten, Conv2D, MaxPool2D
from keras.optimizers import Adam, RMSprop
import optuna
# Load the data
data = pd.read_csv("../data/train.csv")
# first column is the target, the rest of the columns
# are the pixels of the image
# each row is 1 image
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
data.drop(['label'], axis=1), # the images
data['label'], # the target
test_size = 0.1,
random_state=0)
# Re-scale the data
# 255 is the maximum value a pixel can take
X_train = X_train / 255
X_test = X_test / 255
# Reshape image in 3 dimensions:
# height: 28px X width: 28px X channel: 1
X_train = X_train.values.reshape(-1,28,28,1)
X_test = X_test.values.reshape(-1,28,28,1)
# For Keras, we need to create 10 dummy variables,
# one for each digit
# Encode labels to one hot vectors (ex : digit 2 -> [0,0,1,0,0,0,0,0,0,0])
y_train = to_categorical(y_train, num_classes = 10)
y_test = to_categorical(y_test, num_classes = 10)
# Some image examples
g = plt.imshow(X_train[0][:,:,0])
# we will save the model with this name
path_best_model = 'cnn_model.h5'
# starting point for the optimization
best_accuracy = 0
# function to create the CNN
def objective(trial):
# Start construction of a Keras Sequential model.
model = Sequential()
# Convolutional layers.
# We add the different number of conv layers in the following loop:
num_conv_layers = trial.suggest_int('num_conv_layers', 1, 3)
for i in range(num_conv_layers):
# NOTE: As per the below configuration, the parameters of each
# convolutional layer will be identical, even though we draw their
# values every loop; this is achieved with the name: in each trial,
# the value of each hyperparameter defined with a name is identical.
# If we want to have different values in each loop,
# we should change the name, e.g.:
# filters=trial.suggest_categorical(f'filters_{i}', [16, 32, 64])
# kernel_size=trial.suggest_categorical(f'kernel_size_{i}', [3, 5])
# ...
# if we want different parameters in each layer, check next
# notebook
model.add(Conv2D(
filters=trial.suggest_categorical('filters', [16, 32, 64]),
kernel_size=trial.suggest_categorical('kernel_size', [3, 5]),
strides=trial.suggest_categorical('strides', [1, 2]),
activation=trial.suggest_categorical(
'activation', ['relu', 'tanh']),
padding='same',
))
# we could also optimize these parameters if we wanted:
model.add(MaxPool2D(pool_size=2, strides=2))
# Flatten the 4-rank output of the convolutional layers
# to 2-rank that can be input to a fully-connected Dense layer.
model.add(Flatten())
# Add fully-connected Dense layers.
# The number of layers is a hyper-parameter we want to optimize.
# We add the different number of layers in the following loop:
num_dense_layers = trial.suggest_int('num_dense_layers', 1, 3)
for i in range(num_dense_layers):
# Add the dense fully-connected layer to the model.
# This has two hyper-parameters we want to optimize:
# The number of nodes (neurons) and the activation function.
model.add(Dense(
units=trial.suggest_int('units', 5, 512),
activation=trial.suggest_categorical(
'activation', ['relu', 'tanh']),
))
# Last fully-connected dense layer with softmax-activation
# for use in classification.
model.add(Dense(10, activation='softmax'))
# Use the Adam method for training the network.
optimizer_name = trial.suggest_categorical(
'optimizer_name', ['Adam', 'RMSprop'])
if optimizer_name == 'Adam':
optimizer = Adam(lr=trial.suggest_float('learning_rate', 1e-6, 1e-2))
else:
optimizer = RMSprop(
lr=trial.suggest_float('learning_rate', 1e-6, 1e-2),
momentum=trial.suggest_float('momentum', 0.1, 0.9),
)
# In Keras we need to compile the model so it can be trained.
model.compile(optimizer=optimizer,
loss='categorical_crossentropy',
metrics=['accuracy'])
# train the model
# we use 3 epochs to be able to run the notebook in a "reasonable"
# time. If we increase the epochs, we will have better performance
# this could be another parameter to optimize in fact.
history = model.fit(
x=X_train,
y=y_train,
epochs=3,
batch_size=128, # this could be another hyperparameter
validation_split=0.1,
)
# Get the classification accuracy on the validation-set
# after the last training-epoch.
accuracy = history.history['val_accuracy'][-1]
# Save the model if it improves on the best-found performance.
# We use the global keyword so we update the variable outside
# of this function.
global best_accuracy
# If the classification accuracy of the saved model is improved ...
if accuracy > best_accuracy:
# Save the new model to harddisk.
# Training CNNs is costly, so we want to avoid having to re-train
# the network with the best found parameters. We save it instead
# as we search for the best hyperparam space.
model.save(path_best_model)
# Update the classification accuracy.
best_accuracy = accuracy
# Delete the Keras model with these hyper-parameters from memory.
del model
# Remember that Scikit-optimize always minimizes the objective
# function, so we need to negate the accuracy (because we want
# the maximum accuracy)
return accuracy
# we need this to store the search
# we will use it in the following notebook
# NOTE: if the SQLite database exists, it is loaded
# and the optimization keeps going from the last trial,
# i.e., all the stored trail history is used.
study_name = "cnn_study" # unique identifier of the study.
storage_name = "sqlite:///{}.db".format(study_name)
study = optuna.create_study(
direction='maximize',
study_name=study_name,
storage=storage_name,
load_if_exists=True,
)
study.optimize(objective, n_trials=10) # 30
study.best_params
study.best_value
results = study.trials_dataframe()
results['value'].sort_values().reset_index(drop=True).plot()
plt.title('Convergence plot')
plt.xlabel('Iteration')
plt.ylabel('Accuracy')
# load best model
model = load_model(path_best_model)
model.summary()
# make predictions in test set
result = model.evaluate(x=X_test,
y=y_test)
# print evaluation metrics
for name, value in zip(model.metrics_names, result):
print(name, value)
# Predict the values from the validation dataset
y_pred = model.predict(X_test)
# Convert predictions classes to one hot vectors
y_pred_classes = np.argmax(y_pred, axis = 1)
# Convert validation observations to one hot vectors
y_true = np.argmax(y_test, axis = 1)
# compute the confusion matrix
cm = confusion_matrix(y_true, y_pred_classes)
# let's make it more colourful
classes = 10
plt.imshow(cm, interpolation='nearest', cmap=plt.cm.Blues)
plt.title('Confusion matrix')
plt.colorbar()
tick_marks = np.arange(classes)
plt.xticks(tick_marks, range(classes), rotation=45)
plt.yticks(tick_marks, range(classes))
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, cm[i, j],
horizontalalignment="center",
color="white" if cm[i, j] > 100 else "black",
)
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')Notebook: 05-Evaluating-the-search.ipynb.
Visualization backends with same endpoints/functions:
More information: Optuna Visualization
optuna.visualization.matplotlib.plot_contour: Plot the parameter relationship as contour plot in a study with Matplotlib.optuna.visualization.matplotlib.plot_edf: Plot the objective value EDF (empirical distribution function) of a study with Matplotlib.optuna.visualization.matplotlib.plot_intermediate_values: Plot intermediate values of all trials in a study with Matplotlib.optuna.visualization.matplotlib.plot_optimization_history: Plot optimization history of all trials in a study with Matplotlib.optuna.visualization.matplotlib.plot_parallel_coordinate: Plot the high-dimensional parameter relationships in a study with Matplotlib.optuna.visualization.matplotlib.plot_param_importances: Plot hyperparameter importances with Matplotlib.optuna.visualization.matplotlib.plot_pareto_front: Plot the Pareto front of a study.optuna.visualization.matplotlib.plot_slice: Plot the parameter relationship as slice plot in a study with Matplotlib.optuna.visualization.matplotlib.is_available: Returns whether visualization with Matplotlib is available or not.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import optuna
fig = optuna.visualization.matplotlib.plot_optimization_history(study)
optuna.visualization.matplotlib.plot_contour(
study,
params=["num_conv_layers", "num_dense_layers", "optimizer_name", 'units'],
)
optuna.visualization.matplotlib.plot_slice(
study,
params=["num_conv_layers", "num_dense_layers", "optimizer_name", 'units'],
)
optuna.visualization.matplotlib.plot_param_importances(study)
optuna.visualization.matplotlib.plot_parallel_coordinate(
study,
params=["num_conv_layers", "num_dense_layers", "optimizer_name", 'units'],
)
optuna.visualization.matplotlib.plot_edf([study])
# We can even load two studies and plot them together
study_name2 = "cnn_study_2"
storage_name2 = "sqlite:///{}.db".format(study_name2)
study2 = optuna.load_study(
study_name=study_name2,
storage=storage_name2,
)
optuna.visualization.matplotlib.plot_edf([study, study2])There is also an Optuna Dashboard, where we load the study/experiments database and create the plots interactively.
pip install optuna-dashboard
optuna-dashboard sqlite:///example-study.dbThis framework is new: it wasn't mentioned until now. In fact, this section was not in the original course and it was added by me; it introduces Ax, the Adaptive Experimentation Platform.
There is a custom README.md in the folder Section-13-Ax/, along with a custom file of requirements.