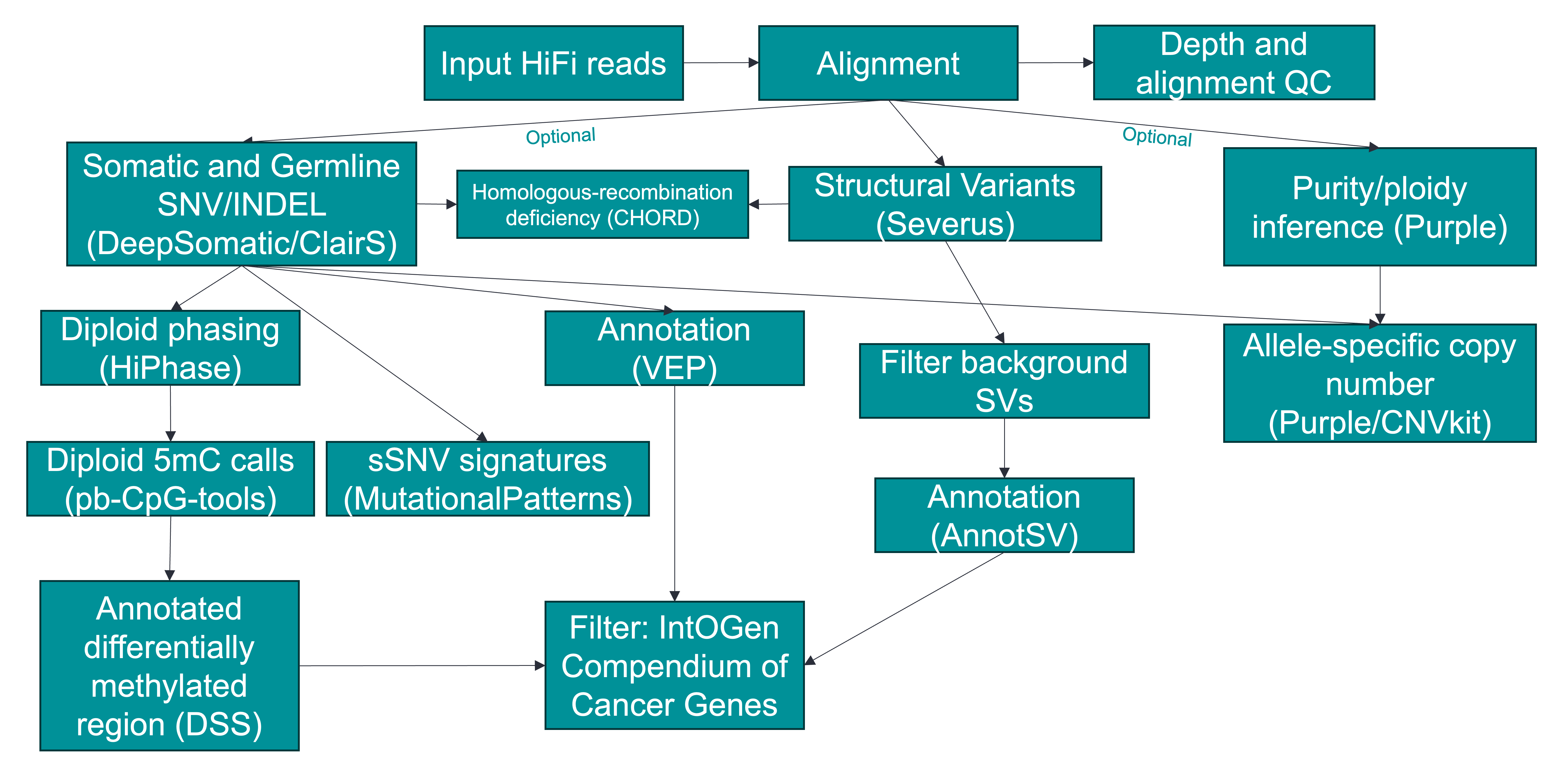
A tumor-normal variant-calling workflow for HiFi data
- Table of contents
- Installation and Dependencies
- Usage
- Important outputs from workflow
- Demo datasets and accuracy of the workflow
- References
- Tools versions
- Change logs
- DISCLAIMER
The workflow is written in WDL version 1.0 (Workflow Description Language). It depends on miniwdl and singularity (version 3 and later). miniwdl can be installed using Bioconda. The workflow can also be run using Cromwell. (See here for instructions, tested on version 86).
A step-by-step tutorial demonstrating usage and an FAQ can be found here.
The workflow will generate the following (non-exhaustive list) results in the $OUTDIR/_LAST/out folder. Please refer to output for a more detailed description of the outputs. An example of final HTML report from COLO829 dataset can be found here (Right click to save the file and double click to open it in a web browser).
| Folder | Types of results |
|---|---|
| AnnotatedSeverusSV | Severus structural variants annotated with AnnotSV (TSV, see AnnotSV README) |
| Annotated*SV_intogen | Structural variants annotated with AnnotSV (TSV) overlapping with the Compendium of Cancer Genes (IntOGen May 23) |
| small_variant_vcf_annotated | DeepSomatic or ClairS SNV/INDEL annotated with VEP (VCF, single entry per variant with --pick, see VEP documentation) |
| small_variant_tsv_annotated | VEP annotation for SNV/INDEL in TSV format |
| small_variant_tsv_CCG | SNV/INDEL that are in the Compendium of Cancer Genes (IntOGen May 23) |
| mutsig_SNV_profile | Mutational profile plot (MutationalPattern) |
| mutsig_SNV | Mutational signature in TSV format (MutationalPattern) |
| normal_germline_small_variant_vcf_annotated | ClairS/Clair3 germline SNV/INDEL (In normal sample) annotated with VEP (Optional, see input JSON parameters) |
| DMR_annotated | Differentially methylated region annotated with genes/introns/promoters etc (TSV) |
| DMR_results | Raw differentially methylated region from DSS (Unannotated, TSV) |
| DMR_annotated_CCG | Annotated DMR (>50 CpG sites) overlapping with the Compendium of Cancer Genes (IntOGen May 23) |
| mosdepth_normal_summary | Depth of coverage of normal (TXT) |
| mosdepth_tumor_summary | Depth of coverage of tumor (TXT) |
| normal_bams_phased | Phased normal BAM file (Hiphase) |
| tumor_bams_hiphase | Phased tumor BAM file (Hiphase) |
| tumor_bams_longphase | Phased tumor BAM file (Optionally, use Longphase for phasing. See input JSON parameters) |
| overall_(tumor|normal)_alignment_stats | Alignment overall statistics (Mapped %) |
| per_alignment_(tumor|normal)_stats | Statistics (accuracy/n_mismatches/length) for each alignment |
| aligned_RL_summary_(tumor|normal) | Aligned read length N50, mean and median |
| normal_germline_small_variant_vcf | Germline variants in normal (VCF) |
| tumor_germline_small_variant_vcf | Germline variants in tumor (VCF) |
| pileup_(normal/tumor)_bed | Summarized 5mC probability in normal and tumor (BED, see pb-CpG-tools for format description) |
| cnvkit_cns_with_major_minor_CN | Copy number segments adjusted with purity and ploidy estimate, see cnvkit_output for raw CNVKit result (BED) |
| Severus_filtered_vcf | Severus structural variants (filtered with control VCF and has simple annotation based on svpack) |
| small_variant_vcf | DeepSomatic or ClairS SNV/INDEL (Unannotated VCF) |
| Purple_outputs | Purity and ploidy estimate + allele-specific copy number calls from HMFtools suite |
| chord_hrd_prediction | Homologous recombination deficiency (HRD) prediction using CHORD |
| report | HTML report summarizing the results. This can be open in any modern web browser. The report is only generated if all steps in the pipeline is carried out (e.g. small variants calling, SV annotation) |
There are two cancer cell lines sequenced on Revio systems, provided by PacBio:
- COLO829 (60X tumor, 60X normal): https://downloads.pacbcloud.com/public/revio/2023Q2/COLO829
- HCC1395 (60X tumor, 40X normal): https://downloads.pacbcloud.com/public/revio/2023Q2/HCC1395/
A benchmark of both cell lines can be found here.
References of tools used
Following are the references for the tools used in the workflow, which should be cited if you use the workflow. The list may not be exhaustive; we welcome suggestions for additional references.
- Zheng, Z. et al. ClairS: a deep-learning method for long-read somatic small variant calling. 2023.08.17.553778 Preprint at https://doi.org/10.1101/2023.08.17.553778 (2023).
- English, A. C., Menon, V. K., Gibbs, R. A., Metcalf, G. A. & Sedlazeck, F. J. Truvari: refined structural variant comparison preserves allelic diversity. Genome Biology 23, 271 (2022).
- Wang, T. et al. The Human Pangenome Project: a global resource to map genomic diversity. Nature 604, 437–446 (2022).
- Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
- Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
- Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single molecule sequencing. Nat Methods 15, 461–468 (2018).
- Pedersen, B. S. & Quinlan, A. R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018).
- McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biology 17, 122 (2016).
- Park, Y. & Wu, H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 32, 1446–1453 (2016).
- Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 12, e1004873 (2016).
- Martínez-Jiménez, F. et al. A compendium of mutational cancer driver genes. Nat Rev Cancer 20, 555–572 (2020). https://www.intogen.org
- Manders, F. et al. MutationalPatterns: the one stop shop for the analysis of mutational processes. BMC Genomics 23, 134 (2022).
- Lin, J.-H., Chen, L.-C., Yu, S.-C. & Huang, Y.-T. LongPhase: an ultra-fast chromosome-scale phasing algorithm for small and large variants. Bioinformatics 38, 1816–1822 (2022).
- HMFtools suite (Amber, Cobalt and Purple): https://github.com/hartwigmedical/hmftools/tree/master
Tools used in the workflow
| Tool | Version | Purpose |
|---|---|---|
| pbmm2 | 1.12.0 | Alignment of HiFi reads |
| pbtk | 3.1.0 | Merging HiFi reads |
| samtools | 1.17 | Various tasks manipulating BAM files |
| VEP | 110.1 | Annotation of small variants |
| AnnotSV | 3.3.6 | Annotation of structural variants |
| DSS | 2.48.0 | Differential methylation |
| annotatr | 1.26.0 | Annotation of differentially methylated region (DMR) |
| ClairS | 0.1.6 | Somatic SNV and INDEL caller |
| bcftools | 1.17 | Manipulation of VCF |
| CNVKit | 0.9.10 | Copy number segmentation |
| Truvari | 4.0.0 | Filtering of control structural variants (Deprecated, using svpack instead) |
| bedtools | 2.31.0 | Splitting genome intervals for parallelization |
| mosdepth | 0.3.4 | Calculating depth of coverage |
| pb-CpG-tools | 2.3.1 | Summarizing 5mC probability |
| HiPhase | 1.1.0 | Diploid phasing using germline variants |
| slivar | 0.3.0 | Selecting/filtering variants from VCF |
| Severus | v0.1.1 | Structural variants |
| seqkit | 2.5.1 | Aligned BAM statistics |
| csvtk | 0.27.2 | Aligned BAM statistics summary and other CSV/TSV operation |
| IntOGen | May 31 2023 | Compendium of Cancer Genes for annotation |
| MutationalPattern | 3.10.0 | Mutational signatures based on SNV |
| Longphase | v1.5.2 | Optional phasing tool |
| Amber | v4.0 | BAF segmentation (HMFtools suite) |
| Cobalt | v1.16.0 | Log ratio segmentation (HMFtools suite) |
| Purple | v4.0 | Purity and ploidy estimate, somatic CNV (HMFtools suite) |
| DeepSomatic | v1.6.1 | Somatic SNV/INDELs caller |
| CHORD | v2.0.0 | HRD prediction |
Click to expand changelogs:
-
v0.7:
- Updated DeepSomatic to v1.6.1.
- Pipeline now calls DeepSomatic in chunks.
-
v0.6.2:
- Added experimental CHORD HRD (Homologous Recombination Deficiency) prediction. See here for details.
- Renamed some variables (legacy Sniffles parameters).
- Swap CNVkit visualization to Purple in report.
- Made changes in WDL for compatibility with Cromwell.
- Allow specifying min/max purity/ploidy for Purple (In task WDL)
-
v0.6.1:
- Updated documentation and benchmark.
- Small bugfix in tabix indexing of VCFs.
- Added HTML report for summary metrics.
- Fixed a bug where germline VCFs are not output when using DeepSomatic.
- Fixed a bug introduced in svpack filtering where entries with
SVLEN=0. are filtered out (They should not be).
-
v0.6:
- Added DeepSomatic 1.6.0 (Experimental and disabled by default. Enable with
hifisomatic.use_deepsomaticin input JSON). Note that DeepSomatic is computationally expensive compared to ClairS so we recommend disabling it if computational resources are limited. See benchmark here for comparison between ClairS and DeepSomatic. - Use
svpackto filter for control SVs (previously using truvari) and provide simple annotation in filtered VCF. - Switch to using
samtoolsto strip kinetics.
- Added DeepSomatic 1.6.0 (Experimental and disabled by default. Enable with
-
v0.5:
- Updated Cobalt to 4.0. It now counts read depth correctly. See here for details.
- Containers are now on pacbio quay.io.
- SV calling now only uses Severus.
- Individual tasks now output the version number in stdout.
-
v0.4:
- Added purity, ploidy and somatic CNV with Amber, Cobalt and Purple
- Note that Cobalt doesn't count the read depth from long-reads correctly so it'll affect the segmentation accuracy. However, purity and ploidy estimation appears to be robust.
- CNVKit segmentation results recalled with purity and ploidy estimate from Purple.
- Severus release now updated with Bioconda container.
- Fixed an issue when call_smallvariants is set to false (Issue #1).
- Added purity, ploidy and somatic CNV with Amber, Cobalt and Purple
-
v0.3:
- Added IntOGen filtering of SV/SNV/INDEL/DMR.
- Added mutational signature analysis.
- Added germline small variants annotation with VEP (optional).
- Added Longphase as an optional phasing tool.
- Better documentation of output in output.
-
v0.2:
- Downgraded to WDL 1.0 for better compatibility.
- Added run time attribute to tasks for future support on cloud (not tested yet).
-
v0.1: Initial release.
TO THE GREATEST EXTENT PERMITTED BY APPLICABLE LAW, THIS WEBSITE AND ITS CONTENT, INCLUDING ALL SOFTWARE, SOFTWARE CODE, SITE-RELATED SERVICES, AND DATA, ARE PROVIDED "AS IS," WITH ALL FAULTS, WITH NO REPRESENTATIONS OR WARRANTIES OF ANY KIND, EITHER EXPRESS OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, ANY WARRANTIES OF MERCHANTABILITY, SATISFACTORY QUALITY, NON-INFRINGEMENT OR FITNESS FOR A PARTICULAR PURPOSE. ALL WARRANTIES ARE REJECTED AND DISCLAIMED. YOU ASSUME TOTAL RESPONSIBILITY AND RISK FOR YOUR USE OF THE FOREGOING. PACBIO IS NOT OBLIGATED TO PROVIDE ANY SUPPORT FOR ANY OF THE FOREGOING, AND ANY SUPPORT PACBIO DOES PROVIDE IS SIMILARLY PROVIDED WITHOUT REPRESENTATION OR WARRANTY OF ANY KIND. NO ORAL OR WRITTEN INFORMATION OR ADVICE SHALL CREATE A REPRESENTATION OR WARRANTY OF ANY KIND. ANY REFERENCES TO SPECIFIC PRODUCTS OR SERVICES ON THE WEBSITES DO NOT CONSTITUTE OR IMPLY A RECOMMENDATION OR ENDORSEMENT BY PACBIO.