Note: The master branch is currently developing the 2.0 version of BAT. For the latest stable version download the package from the BATv1.0 branch.
See also: GHOAT.py, a fully automated tool for guest-host ABFE calculations using SDR with pmemd.cuda:
https://github.com/GHeinzelmann/GHOAT.py
A tutorial and a detailed user guide are available, as well as necessary parameter and input files for several hosts.
The Binding Affinity Tool (BAT.py) is a python tool for fully automated absolute binding free energy calculations. Its workflow encompasses the creation of the bound complex, generation of parameters using Antechamber, preparation of the simulation files, and post-processing to retrieve the binding free energy. By using the pmemd.cuda software from AMBER20, it is able to perform several calculations at a reduced computational cost using graphics processing units (GPUs).
BAT.py can perform binding free energy calculations by two alchemical routes in the presence of restraints, either with double decoupling (DD) procedure or with the simultaneous decoupling recoupling (SDR) method. It can also apply a physical route, through the APR method, suitable for ligands that have free access to the solvent. The program is compatible with the simulation package AMBER20, also requiring a few installed programs to work properly, which are listed in the next section.
To use BAT.py, download the files from this repository, which already contain an example for ligand binding to the second bromodomain of the BRD4 protein - BRD4(2). In order to perform all the steps from BAT.py, the following programs must be installed and in your path:
VMD (Visual Molecular Dynamics) [1] - https://www.ks.uiuc.edu/Development/Download/download.cgi?PackageName=VMD
Openbabel 2.4.1 [2] - https://github.com/openbabel/openbabel/releases/tag/openbabel-2-4-1 †
MUSTANG v3.2.3 (MUltiple (protein) STructural AligNment alGorithm) [3] - http://lcb.infotech.monash.edu.au/mustang/
AmberTools20 or later [4] - http://ambermd.org/AmberTools.php ‡
pmemd.cuda software from AMBER20 [4] - http://ambermd.org/GetAmber.php
† Had problems with wrong protonation using Openbabel 3, so keeping the 2.4.1 version for now, might change in the future.
‡ If pdb4amber from Ambertools does not work, add the following line to your amber.sh file: export PYTHONPATH=$PYTHONPATH:$AMBERHOME/lib/python3.8/site-packages/pdb4amber-1.7.dev0-py3.8.egg/
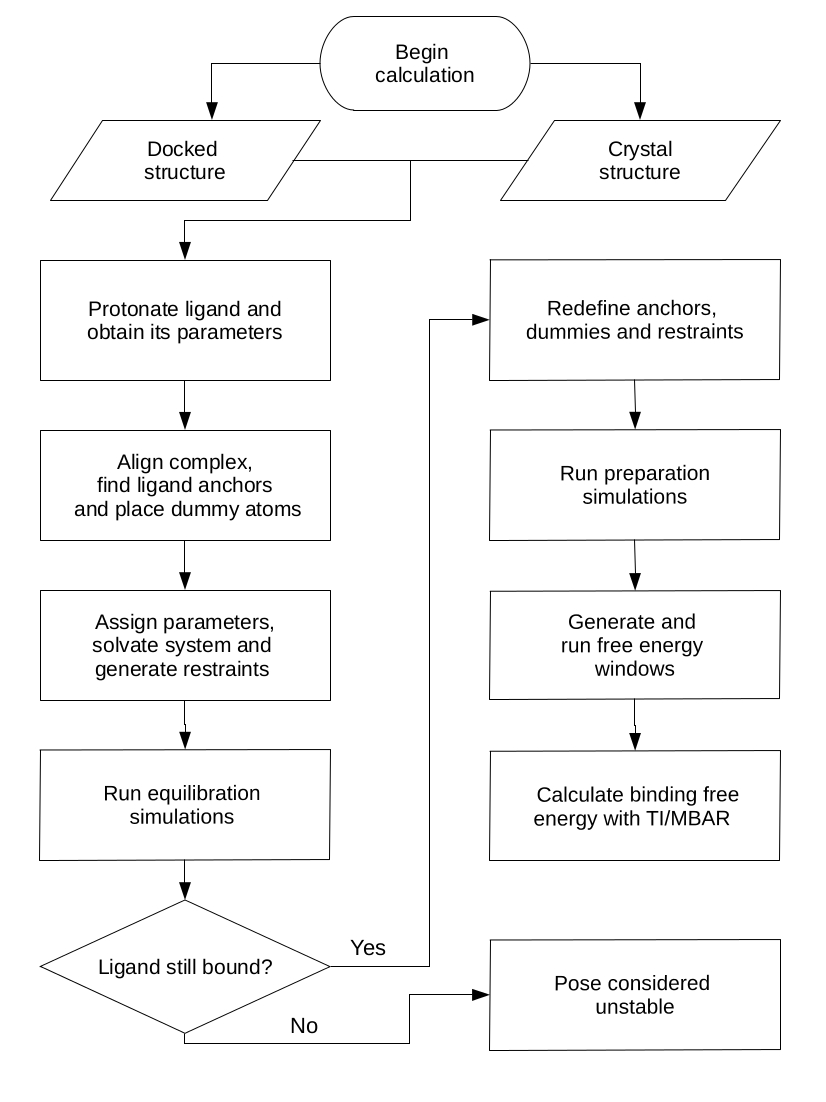
The folder ./BAT/all-poses contains an example of system input files, with a docked receptor from the 5uez crystal structure (LMCSS-5uf0_5uez_docked.pdb), as well as 9 docked poses for the ligand with the 5uf0 crystal structure (pose0.pdb to pose8.pdb). The docking files were generated and converted to .pdb using Autodock Vina and AutodockTools, following a protocol adapted from the CELPP challenge tutorial (https://docs.google.com/document/d/1iJcPUktbdrRftAA8cuVa32Ri1TPr2XvZVqTccDja2OM). Inside the ./all-poses folder there is also the original crystal structure file for 5uf0. Below we show an example of using these files to calculate the standard binding free energies of the top 5 docked poses and the crystal structure, with all the necessary steps in the calculation.
The simulations and analysis from this example will be performed inside the ./BAT folder. They are divided in three steps, equilibration (folder ./equil), preparation (folder ./prep) and free energy calculation (folder ./fe). The input file with all the needed BAT.py parameters for double decoupling is called input-dd.in, with the meaning of each explained in more detail in the user guide, located inside the ./doc folder. For our sample calculation, we will use the values already provided in the input files included in this distribution. Briefly, the poses_list parameter sets up the calculation for the first 5 poses from Autodock Vina, all in the ./all-poses folder. The input files can be modified to perform the calculations in the 5uf0 crystal structure, by changing the calc_type option to "crystal", the celpp_receptor option to "5uf0", and the ligand_name option to "89J", which is the ligand residue name in the 5uf0 pdb structure.
The equilibration step starts from the docked complex or the crystal structure, gradually releasing restraints applied on the ligand and then performing a final simulation with an unrestrained ligand. The necessary simulation parameters for the ligand are also generated in this stage, using the General Amber Force Field versions 1 and 2 (GAFF or GAFF2) [5], and the AM1-BCC charge model [6,7]. To run this step, inside the program main folder type:
python BAT.py -i input-dd.in -s equil
BAT.py is compatible with python 3.8 versions. If you have another version, or you find that this command gives an error, you can use the python version included in the Ambertools20 distribution:
$AMBERHOME/miniconda/bin/python BAT.py -i input-dd.in -s equil
This command will create an ./equil folder, with one folder inside for each of the docked poses (pose0, pose1, etc.). In order to run the simulations for each pose, you can use the run-local.bash script (to run them locally), or the PBS-run script, which is designed to run in a queue system such as TORQUE. Both of these files might have to be adjusted, depending on your computer or server configuration. The number of simulations and the applied restraints will depend on the release_eq array defined in the input file.
The second stage starts from the equilibrated system, rebuilding the latter, as well as redefining the dummy/anchor atoms and the restraints for use in the free energy calculation. To run this stage, type in the program main folder:
python BAT.py -i input-dd.in -s prep
, or use the miniconda option as shown in the previous section. It is possible that the ligand has left the binding site during equilibration, due to an unstable docked pose. In this case, the preparation is not performed for this pose, and a warning message appears after running the command above. The same way as the equil stage, there is a folder for each pose (given that it did not unbind during equilibration) in the ./prep directory, and the simulations can be run locally or in a queue system such as TORQUE. Once the preparation simulations are concluded, the systems are ready for the binding free energy calculations.
Starting from the states created in the prep stage, we can now perform the binding free energy calculations, which will be located inside the ./fe folder. In this example we will use the double decoupling method (DD) with retraints to obtain the binding free energies. For charged ligands, one should use the simultaneous decoupling recoupling (SDR) method instead, as explained in the user guide. Again in the program main folder, type:
python BAT.py -i input-dd.in -s fe
For each pose or crystal structure, a folder will be created inside ./fe, and inside there will be two folders, ./restraints and ./dd. The restraints folder contains all the simulations needed for the application/removal of restraints. The ./dd folder contains the coupling/decoupling of the ligand electrostatic/LJ interactions, both in the binding site and in bulk. A script called run-all.bash, inside the ./run_files folder, can be used to run these simulatons quickly using the PBS scripts provided. A similar script can be written to do the same, using your particular running protocol.
Once all of the simulations are concluded, it is time to process the output files and obtain the binding free energies. Here a few parameters concerning the analysis can be set in the input file, such as using TI or MBAR [8] for double decoupling, number of blocks for block data analysis, and the Gaussian weights if TI is used for double decoupling. Inside the main folder type:
python BAT.py -i input-dd.in -s analysis
You should see a ./Results directory inside each ./fe/pose folder, containing all the components and the final calculated binding free energy, located in the Results.dat file. This folder also contains the results for each of the chosen data blocks, which is useful to check for convergence and fluctuations, and is also used to calculate the uncertainties. This fully automated procedure can be readily applied for any other ligand that binds to the second BRD4 bromodomain, and with minimal adjustments it can be extended to several other proteins.
The SDR method is suitable for ligands with net charge, since it keeps the full system neutral during the transformations. The APR method presents limitations, but it could stil be useful for ligands that bind to the surface of proteins and have clear access to the solvent [9]. To apply SDR/APR in addition to DD, a few parameters have to be changed or added, which is already demonstrated in the input-sdr.in and input-apr.in input files included in this example.
In the case of APR, the preparation, free energy and analysis steps have to be ran again using the input-apr.in input file. In the case of SDR, only the free energy and analysis stages have to be performed again using the input-sdr.in file. The three methods, especially DD and SDR, should produce consistent results if the reference state is the same from the preparation stage.
The sample system shown here uses a particular ligand that binds to the second bromodomain of the BRD4 protein - BRD4(2). The system alignment, parameter generation, assignment of the ligand anchor atoms, and positioning of the dummy atoms is done automatically, so these same calculations can be extended to any other ligand that binds to this receptor. The only thing needed is the files in the ./all-poses folder to be changed, including the docked receptor and poses pdb files, as well as the crystal structure if desired.
To include a new receptor system, some additional input data is needed. They include a reference.pdb file to align the system using MUSTANG, three chosen protein anchors, and a few variables for ligand anchor atom search. These can be found inside the ./systems-library folder for three other bromodomains (CREBBP, BRD4(1) and BAZ2B), T4 Lysozyme, the Major Urinary Protein (MUP), and the Retinol binding protein 1 (RetBP). Other systems will be added with time, as the program is further tested and validated.
A paper explaining the whole BAT.py theoretical aspects and calculation procedure is available in Ref [9]. For more information you can contact the author directly:
Germano Heinzelmann
Departamento de Física, Universidade Federal de Santa Catarina
Florianópolis - SC 88040-970 Brasil
email: germanohei@gmail.com
Germano Heinzelmann thanks FAPESC and CNPq for the research grants.
-
W. Humphrey, A. Dalke and K. Schulten. (1996) "VMD - Visual Molecular Dynamics", Journal of Molecular Graphics, 14, 33-38.
-
N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch, and G. R. HutchisonEmail. (2011) "Open Babel: An open chemical toolbox." Journal of Cheminformatics, 3, 33.
-
A. S. Konagurthu, J. Whisstock, P. J. Stuckey, and A. M. Lesk. (2006) “MUSTANG: A multiple structural alignment algorithm”. Proteins, 64, 559-574.
-
D.A. Case, K. Belfon, I.Y. Ben-Shalom, S.R. Brozell, D.S. Cerutti, T.E. Cheatham, III, V.W.D. Cruzeiro, T.A. Darden, R.E. Duke, G. Giambasu, M.K. Gilson, H. Gohlke, A.W. Goetz, R. Harris, S. Izadi, S.A. Izmailov, K. Kasavajhala, A. Kovalenko, R. Krasny, T. Kurtzman, T.S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, V. Man, K.M. Merz, Y. Miao, O. Mikhailovskii, G. Monard, H. Nguyen, A. Onufriev, F.Pan, S. Pantano, R. Qi, D.R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C. Simmerling, N.R.Skrynnikov, J. Smith, J. Swails, R.C. Walker, J. Wang, L. Wilson, R.M. Wolf, X. Wu, Y. Xiong, Y. Xue, D.M. York and P.A. Kollman (2020), AMBER 2020, University of California, San Francisco.
-
J. Wang, R.M. Wolf, J.W. Caldwell, and P. A. Kollman, D. A. Case (2004) "Development and testing of a general AMBER force field". Journal of Computational Chemistry, 25, 1157-1174.
-
A. Jakalian, B. L. Bush, D. B. Jack, and C.I. Bayly (2000) "Fast, efficient generation of high‐quality atomic charges. AM1‐BCC model: I. Method". Journal of Computational Chemistry, 21, 132-146.
-
A. Jakalian, D. B. Jack, and C.I. Bayly (2002) "Fast, efficient generation of high‐quality atomic charges. AM1‐BCC model: II. Parameterization and validation". Journal of Computational Chemistry, 16, 1623-1641.
-
M. R. Shirts and J. Chodera (2008) “Statistically optimal analysis of samples from multiple equilibrium states.” Journal of Chemical Physics, 129, 129105.
-
G. Heinzelmann and M. K. Gilson (2021). “Automation of absolute protein-ligand binding free energy calculations for docking refinement and compound evaluation”. Scientific Reports, 11, 1116.