SSDraw is a program that generates publication-quality protein secondary structure diagrams from three-dimensional protein structures. To depict relationships between secondary structure and other protein features, diagrams can be colored by conservation score, B-factor, or custom scoring.
SSDraw also has a colab notebook available at https://colab.research.google.com/github/ethanchen1301/SSDraw/blob/main/SSDraw.ipynb (only usable for Chrome)
SSDraw requires the Biopython and matplotlib modules to be installed.
pip install biopython
pip install matplotlib
pip install numpySSDraw also requires DSSP to be installed in order to generate secondary structure annotations:
conda install -c salilab dsspAlternatively, you can install DSSP either through apt-get (sudo apt-get install dssp), or you can follow the instructions on their github page to make a local installation: https://github.com/cmbi/dssp.
If DSSP fails to install, there is a PyTorch implementation of DSSP that you can install as a workaround:
pip install torch
pip install pydssp
pip install nomkl
SSDraw requires 4 arguments:
- --fasta: the file name sequence or alignment file in fasta format.
- --name: the id of the sequence in the fasta file corresponding to your protein of interest.
- --pdb: the file name of the pdb file of your protein
- --output: the output file name to use
python3 ../SSDraw.py --fasta 1ndd.fasta --name 1ndd --pdb 1ndd.pdb --output 1ndd_out
SSDraw uses a gradient to color each position in the alignment by a certain score. The user can choose which scoring system to use, and they can also choose which colormap.
-conservation_score: score each position in the alignment by conservation score.
-bfactor: score each residue in the pdb by B-factor.
-scoring_file: score each residue by a custom scoring file prepared by the user.
-mview: color each residue by the mview coloring system.
python3 ../SSDraw.py --fasta aligned.fasta --name 1ndd --pdb 1ndd.pdb --output 1ndd_conservation -conservation_score --start 80 --end 132
 Note: for more on how the --start and --end options work, see choosing a subregion.
Note: for more on how the --start and --end options work, see choosing a subregion.
python3 ../SSDraw.py --fasta 1ndd.fasta --name 1ndd --pdb 1ndd.pdb --output 1ndd_bfactor -bfactor
The default colormap for SSDraw is inferno. The user can select one of the matplotlib library color maps or simply list a space delimited set of colors they'd like to use with the --color_map option. Alternatively, the user can select a single color with the --color option and SSDraw will use that color on the whole image.
python3 ../SSDraw.py --fasta 2kdl.fasta --name 2kdl --pdb 2kdl.pdb --output 2kdl_out --scoring_file 2kdl_scoring.txt --color_map black cyan
Normally, SSDraw will generate a DSSP annotation from the PDB file, but if you have a DSSP file you would like to use, you can upload it and input the file name in Options.
If you want SSDraw to draw only a portion of your alignment, you can specify the start and/or end points using the --start and --end options respectively. The argument for these options correspond to the index of the alignment position, not to the residue position numbers. See example 2.
We now provide the option to color by ConSurf grade using --consurf option:
python3 ../SSDraw.py --fasta 2n1v.fasta --name 2n1v --pdb 2n1v.pdb --output 2n1v_consurf --consurf 2n1v_nscores.txt --ticks 5
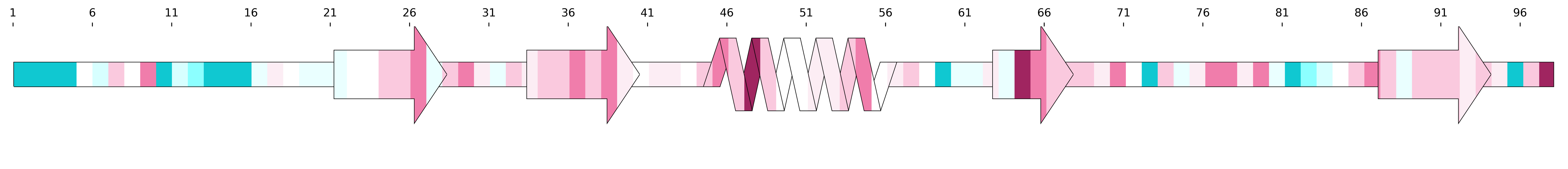 The code can read in either the ConSurf grades file or a Rate4Site file with raw scores. The raw scores will be converted into grades according to the algorithm used by ConSurf: https://github.com/Rostlab/ConSurf
The code can read in either the ConSurf grades file or a Rate4Site file with raw scores. The raw scores will be converted into grades according to the algorithm used by ConSurf: https://github.com/Rostlab/ConSurf
We now provide a helper script to assist the user in stacking multiple diagrams.
python3 run_multiple_pdbs_on_one_msa.py --input [input script] --output [output directory]
run_multiple_pdbs_on_one_msa.py will run SSDraw for multiple pdbs from a single multiple sequence alignment, saving the diagrams to a specified output directory. Finally, the script will create a composite stacked image of the diagrams. An example input script is shown in SSDraw/examples/example_run.txt.