If you use this work, please cite our manuscript: doi
Which epigenetic factors are the best predictors of gene expression? An analysis of histone marks using ROADMAP data.
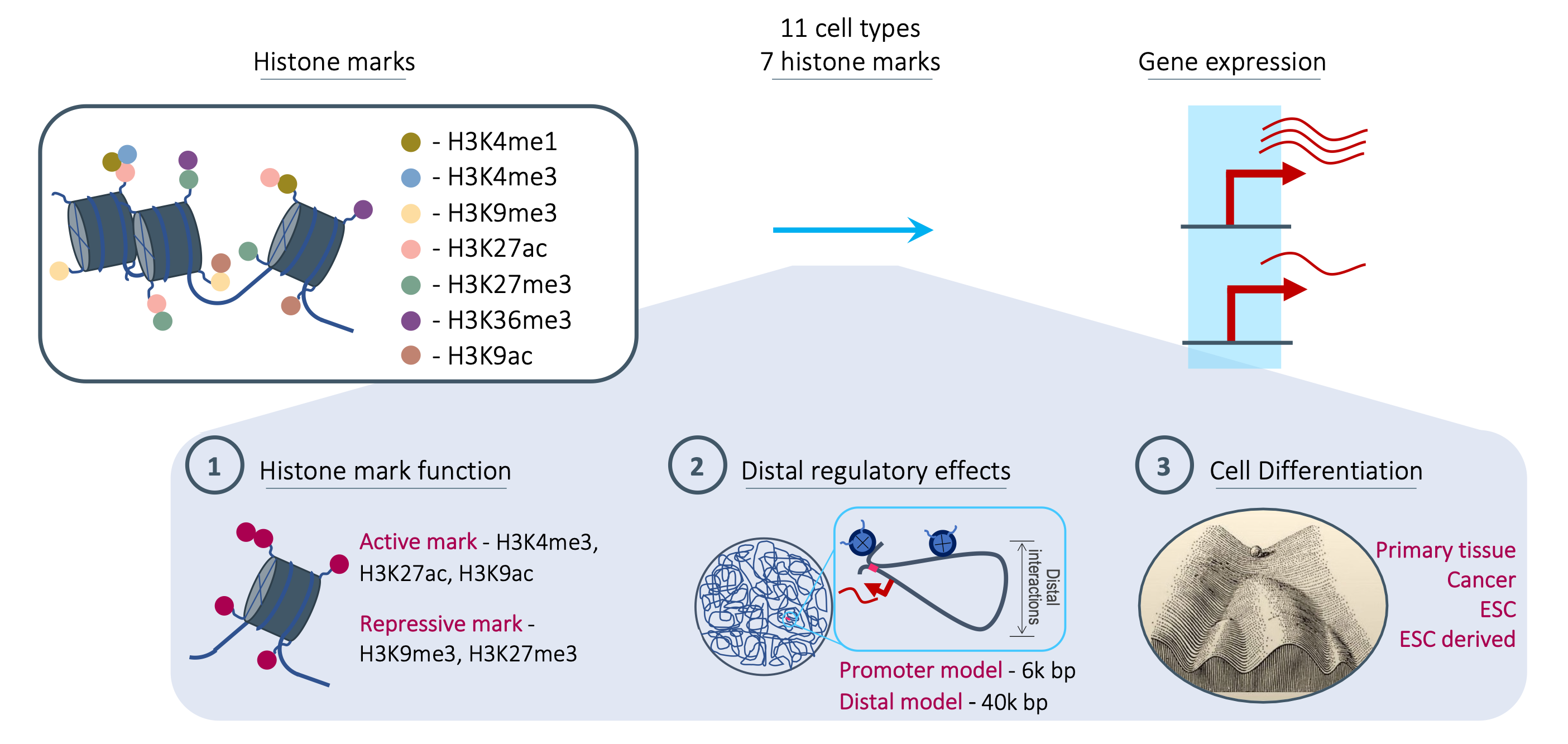
Multiple past approaches have tried to address which histone marks are most predictive of expression but none have considered multiple histone marks, the distance of the regulatory effects and different cell types. Here we address this issue and investigate the effect each plays on the most informative histone marks. See our paper for full details.
Here, we present the most comprehensive study of this relationship to date - Investigating seven histone marks, in eleven cell types, across a diverse range of cell states and utilising both convolutional and attention-based models to account for histone mark activity at promoter regions up to distal regulatory signals. Our work shows how histone mark function, regulatory distance and cellular states collectively influence histone marks’ relationship with transcription. Moreover, we find no universal histone mark which is consistently the most predictive of expression which highlights the need to consider all three of these factors when determining the effect of histone mark activity on the transcriptional state of a cell.
Roadmap data was used, specifically consolidated ChIP-seq read alignments and RPKM expression values for seven major histone marks:
- H3K4me1
- H3K4me3
- H3K9me3
- H3K27me3
- H3K36me3
- H3K27ac
- H3K9ac
This was investigated in eleven cell lines and tissue samples. Note that although the scripts and frameworks differ for the two models, the data, training and testing approach used is the same so comparisons across them are valid.
The results of our work are based on two models a promoter model (ChromExpress - a custom, convolutional neural network (CNN)) and a distal model, chromoformer, transformer-based, DNA interaction-aware deep learning architecture.
We have separate conda enviornments and scripts to run the model training and evaluation for each and have split the repository by model.
Use the conda environments (yaml files in ./environments) for the steps:
conda env create -f ./environments/chromexpress.yml && \
conda env create -f ./environments/chromoformer.yml && \
conda activate chromoformer && \
conda install pytorch==1.9.0 torchvision==0.10.0 torchaudio==0.9.0 torchmetrics=3.0.9 cudatoolkit=11.1 -c pytorch -c conda-forge
Follow steps in chromoformer repository embedded in this
repository (./chromoformer folder) to download all data for model training an evaluation.
Use ./train_run.py or ./train_iter_run.py to train the model for all cell types and histone marks for 4 fold
cross validation. Just pass in the cell and histone mark to train on.
Use ./train_run_chromoformer.py or ./train_iter_run_chromoformer.py to train the model for all cell types and
histone marks for 4 fold cross validation. Note that these scripts can also be used to train on combinations of
histone marks by inputting these as a list.
Use ./bin/test_chromexpress.py to test the model for all cell types and histone marks for 4 fold
cross validation.
Use ./bin/test_chromoformer.py and ./bin/test_chromoformer_combns.py to measure performance of the model for
all cell types and histone marks for 4 fold cross validation. Note that test_chromoformer_combns.py is
specifcally for the model trained to predict expression from two histone marks.
We evaluate the histone mark activity in the receptive field of the models. To rerun this analysis use the following:
Use ./bin/chromexpress_hist_mark_activity.py. Note that this will get histone mark activity for all cell types,
marks and genes not just those in the test set.
Use ./bin/chromoformer_hist_mark_activity.py. Note that this will get histone mark activity for all cell types,
marks and genes not just those in the test set.
See ./bin/In_silico_perturb_chromoformer.py and in_silico_perturb_bootstrap_qtl_enrichment.py for the scripts to analyse the effect on gene expression of degrading activating/repressing histone mark signals at differing distances from the transcriptional start site and looking for enrichment in QTL sets. Analysis of benchmark approaches can be found for max histone mark activity ( in_silico_perturb_bootstrap_max_hist_activity_enrichment.py), proximity (in_silico_perturb_bootstrap_min_dist_enrichment.py) and Hi-C (in_silico_perturb_bootstrap_HiC_enrichment.py).